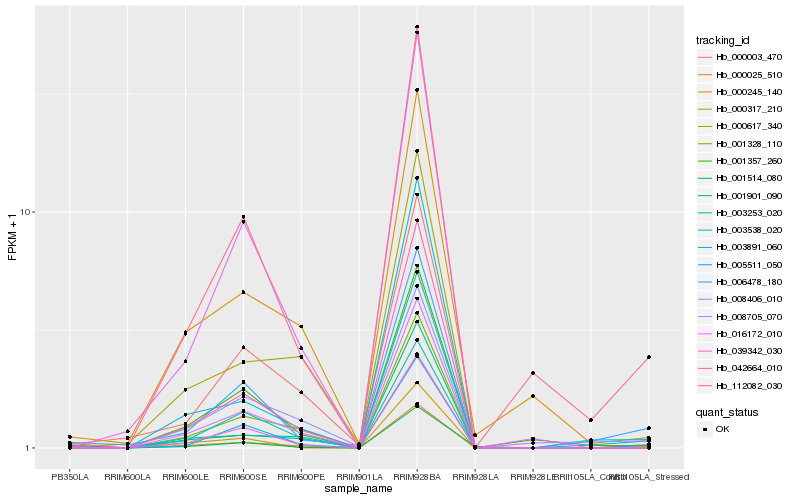
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000025_510 |
0.0 |
- |
- |
PREDICTED: receptor-like protein 12 [Vitis vinifera] |
| 2 |
Hb_006478_180 |
0.1427584617 |
- |
- |
Nonspecific lipid-transfer protein precursor, putative [Ricinus communis] |
| 3 |
Hb_000317_210 |
0.1551773863 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB39 [Jatropha curcas] |
| 4 |
Hb_112082_030 |
0.1561579469 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 5 |
Hb_008705_070 |
0.1568332169 |
- |
- |
PREDICTED: uncharacterized protein LOC105638774 [Jatropha curcas] |
| 6 |
Hb_005511_050 |
0.1596927468 |
- |
- |
hypothetical protein POPTR_0008s04130g [Populus trichocarpa] |
| 7 |
Hb_042664_010 |
0.1612981053 |
- |
- |
hypothetical protein POPTR_0001s07640g, partial [Populus trichocarpa] |
| 8 |
Hb_001514_080 |
0.1640628048 |
- |
- |
hypothetical protein JCGZ_16082 [Jatropha curcas] |
| 9 |
Hb_003538_020 |
0.1648661702 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |
| 10 |
Hb_039342_030 |
0.1663265734 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003253_020 |
0.1665865658 |
transcription factor |
TF Family: B3 |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_001328_110 |
0.1667759018 |
- |
- |
PREDICTED: LON peptidase N-terminal domain and RING finger protein 1-like [Jatropha curcas] |
| 13 |
Hb_000617_340 |
0.1676850849 |
- |
- |
respiratory burst oxidase B [Manihot esculenta] |
| 14 |
Hb_003891_060 |
0.1677012234 |
- |
- |
PREDICTED: linoleate 13S-lipoxygenase 2-1, chloroplastic-like [Glycine max] |
| 15 |
Hb_001901_090 |
0.1687138831 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 16 |
Hb_000245_140 |
0.1699512717 |
- |
- |
PREDICTED: CASP-like protein 3A1 [Jatropha curcas] |
| 17 |
Hb_008406_010 |
0.1701938507 |
- |
- |
PREDICTED: tyrosine aminotransferase-like [Populus euphratica] |
| 18 |
Hb_016172_010 |
0.170746354 |
- |
- |
hypothetical protein JCGZ_23602 [Jatropha curcas] |
| 19 |
Hb_000003_470 |
0.1709130857 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001357_260 |
0.1730734592 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |