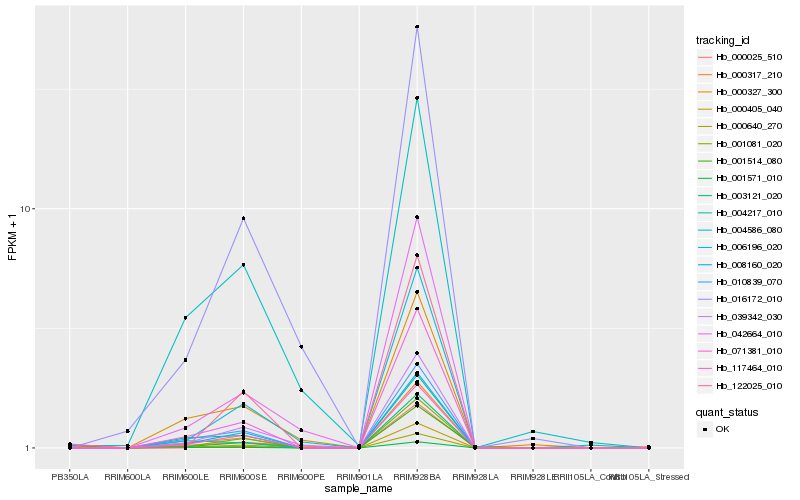
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000317_210 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB39 [Jatropha curcas] |
| 2 |
Hb_071381_010 |
0.1047521802 |
- |
- |
hypothetical protein JCGZ_21650 [Jatropha curcas] |
| 3 |
Hb_010839_070 |
0.1078794694 |
- |
- |
uncharacterized protein LOC100810725 [Glycine max] |
| 4 |
Hb_001571_010 |
0.1126678928 |
- |
- |
hypothetical protein JCGZ_15779 [Jatropha curcas] |
| 5 |
Hb_117464_010 |
0.1133220505 |
- |
- |
PREDICTED: flavone 3'-O-methyltransferase 1-like [Jatropha curcas] |
| 6 |
Hb_008160_020 |
0.1135629362 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 23-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000640_270 |
0.1229272456 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable pectinesterase 30 [Jatropha curcas] |
| 8 |
Hb_001081_020 |
0.1312120373 |
- |
- |
Cytochrome P450 76C2 [Triticum urartu] |
| 9 |
Hb_000405_040 |
0.1449882845 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 10 |
Hb_039342_030 |
0.1462203368 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_006196_020 |
0.1521326766 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 100-like [Populus euphratica] |
| 12 |
Hb_001514_080 |
0.1547977958 |
- |
- |
hypothetical protein JCGZ_16082 [Jatropha curcas] |
| 13 |
Hb_000025_510 |
0.1551773863 |
- |
- |
PREDICTED: receptor-like protein 12 [Vitis vinifera] |
| 14 |
Hb_042664_010 |
0.1553062672 |
- |
- |
hypothetical protein POPTR_0001s07640g, partial [Populus trichocarpa] |
| 15 |
Hb_003121_020 |
0.1593859579 |
- |
- |
secretory peroxidase PX3 [Manihot esculenta] |
| 16 |
Hb_000327_300 |
0.1610802971 |
- |
- |
PREDICTED: ABC transporter G family member 6-like [Jatropha curcas] |
| 17 |
Hb_004586_080 |
0.1650134983 |
- |
- |
hypothetical protein POPTR_0002s10520g [Populus trichocarpa] |
| 18 |
Hb_122025_010 |
0.1654857488 |
- |
- |
hypothetical protein POPTR_0003s052201g, partial [Populus trichocarpa] |
| 19 |
Hb_016172_010 |
0.1714128869 |
- |
- |
hypothetical protein JCGZ_23602 [Jatropha curcas] |
| 20 |
Hb_004217_010 |
0.1725864317 |
- |
- |
putative wall-associated kinase family protein [Populus trichocarpa] |