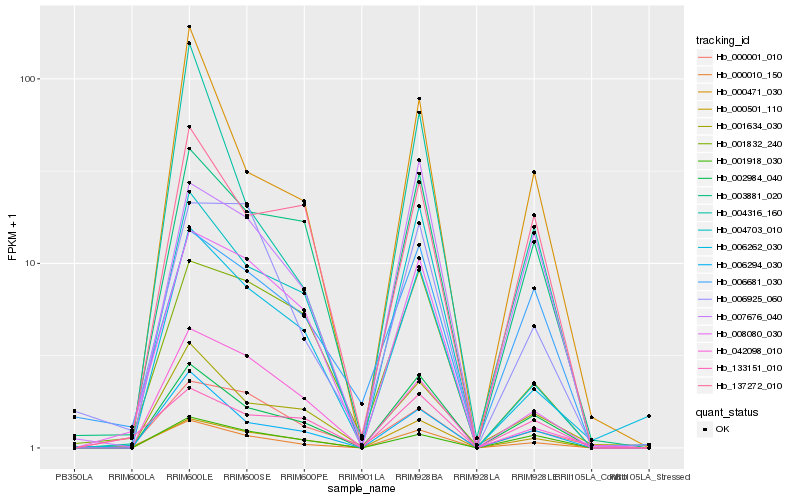
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000010_150 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2-like [Jatropha curcas] |
| 2 |
Hb_002984_040 |
0.1347111368 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_000501_110 |
0.1425215528 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 [Populus euphratica] |
| 4 |
Hb_006294_030 |
0.1450844261 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 5 |
Hb_133151_010 |
0.1656578684 |
- |
- |
NBS-LRR resistance gene-like protein ARGH30 [Populus trichocarpa] |
| 6 |
Hb_000471_030 |
0.1673861978 |
- |
- |
- |
| 7 |
Hb_004703_010 |
0.172451983 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 8 |
Hb_003881_020 |
0.1729949356 |
- |
- |
PREDICTED: stellacyanin-like [Jatropha curcas] |
| 9 |
Hb_006262_030 |
0.1759917308 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 10 |
Hb_006925_060 |
0.1801873508 |
- |
- |
hypothetical protein JCGZ_20651 [Jatropha curcas] |
| 11 |
Hb_001634_030 |
0.1820412872 |
- |
- |
Receptor like protein 1, putative [Theobroma cacao] |
| 12 |
Hb_000001_010 |
0.1823705487 |
- |
- |
PREDICTED: uncharacterized protein LOC105648961 [Jatropha curcas] |
| 13 |
Hb_001918_030 |
0.1850996498 |
- |
- |
- |
| 14 |
Hb_008080_030 |
0.1880285444 |
- |
- |
hypothetical protein POPTR_0275s002101g, partial [Populus trichocarpa] |
| 15 |
Hb_004316_160 |
0.1908282509 |
- |
- |
hypothetical protein CISIN_1g047906mg, partial [Citrus sinensis] |
| 16 |
Hb_006681_030 |
0.1921884733 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X1 [Populus euphratica] |
| 17 |
Hb_007676_040 |
0.1953724512 |
- |
- |
cellulose synthase, putative [Ricinus communis] |
| 18 |
Hb_001832_240 |
0.1966665121 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 19 |
Hb_137272_010 |
0.1983695921 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 10 [Jatropha curcas] |
| 20 |
Hb_042098_010 |
0.1988434432 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |