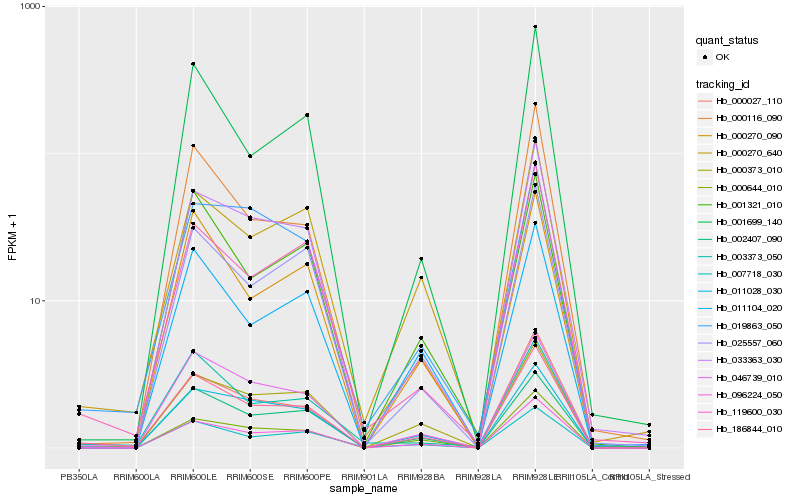
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_186844_010 |
0.0 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 2 |
Hb_007718_030 |
0.090944192 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 3 |
Hb_096224_050 |
0.0920336774 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g31250 [Jatropha curcas] |
| 4 |
Hb_046739_010 |
0.0999967738 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 isoform X1 [Jatropha curcas] |
| 5 |
Hb_033363_030 |
0.101515922 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN7, chloroplastic [Jatropha curcas] |
| 6 |
Hb_002407_090 |
0.1028746741 |
- |
- |
hypothetical protein JCGZ_00747 [Jatropha curcas] |
| 7 |
Hb_000027_110 |
0.1034981172 |
- |
- |
wall-associated kinase, putative [Ricinus communis] |
| 8 |
Hb_119600_030 |
0.1104407046 |
- |
- |
PREDICTED: uncharacterized protein LOC105635187 [Jatropha curcas] |
| 9 |
Hb_001699_140 |
0.1127640163 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000270_640 |
0.1133646533 |
- |
- |
hypothetical protein POPTR_0012s07880g [Populus trichocarpa] |
| 11 |
Hb_000644_010 |
0.113584248 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000270_090 |
0.1150491153 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 13 |
Hb_011028_030 |
0.1164816231 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000116_090 |
0.1166668354 |
- |
- |
catalase [Hevea brasiliensis] |
| 15 |
Hb_025557_060 |
0.117067748 |
transcription factor |
TF Family: MYB |
Myb-related protein 306, putative isoform 1 [Theobroma cacao] |
| 16 |
Hb_001321_010 |
0.118850141 |
- |
- |
HIGH-CHLOROPHYLL-FLUORESCENCE 101 family protein [Populus trichocarpa] |
| 17 |
Hb_011104_020 |
0.1208205504 |
- |
- |
PREDICTED: ABC transporter B family member 2-like [Jatropha curcas] |
| 18 |
Hb_019863_050 |
0.1211395023 |
- |
- |
PREDICTED: uncharacterized protein LOC103343426 [Prunus mume] |
| 19 |
Hb_003373_050 |
0.1215889445 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |
| 20 |
Hb_000373_010 |
0.1220039594 |
- |
- |
ATP binding protein, putative [Ricinus communis] |