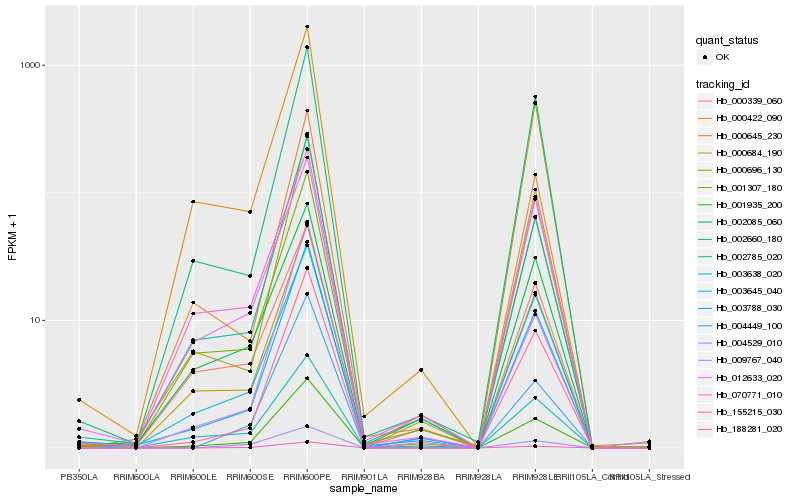
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_155215_030 |
0.0 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 2 |
Hb_004529_010 |
0.0705643188 |
- |
- |
hypothetical protein JCGZ_27055 [Jatropha curcas] |
| 3 |
Hb_001935_200 |
0.0847934346 |
transcription factor |
TF Family: C2H2 |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003788_030 |
0.1115895382 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 5 |
Hb_004449_100 |
0.1260280179 |
- |
- |
- |
| 6 |
Hb_000684_190 |
0.1290205931 |
- |
- |
beta-1,3-glucuronyltransferase, putative [Ricinus communis] |
| 7 |
Hb_003645_040 |
0.1294540893 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0003s13190g [Populus trichocarpa] |
| 8 |
Hb_002785_020 |
0.1295276036 |
- |
- |
PREDICTED: laccase-4-like [Jatropha curcas] |
| 9 |
Hb_009767_040 |
0.1372424924 |
- |
- |
PREDICTED: uncharacterized protein LOC105127022 [Populus euphratica] |
| 10 |
Hb_002085_060 |
0.1407754629 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000645_230 |
0.1431321863 |
- |
- |
PREDICTED: chitinase-like protein 2 [Jatropha curcas] |
| 12 |
Hb_003638_020 |
0.1491394969 |
- |
- |
PREDICTED: uncharacterized protein LOC105645807 [Jatropha curcas] |
| 13 |
Hb_000339_060 |
0.1512866792 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 14 |
Hb_070771_010 |
0.1527909015 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g06890 [Jatropha curcas] |
| 15 |
Hb_001307_180 |
0.1528438725 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 4 [UDP-forming] isoform X1 [Jatropha curcas] |
| 16 |
Hb_012633_020 |
0.153588963 |
- |
- |
laccase, putative [Ricinus communis] |
| 17 |
Hb_000696_130 |
0.1538804763 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 18 |
Hb_188281_020 |
0.1540321922 |
- |
- |
PREDICTED: uncharacterized protein LOC105629522 [Jatropha curcas] |
| 19 |
Hb_000422_090 |
0.1544668746 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002660_180 |
0.1547905917 |
- |
- |
conserved hypothetical protein [Ricinus communis] |