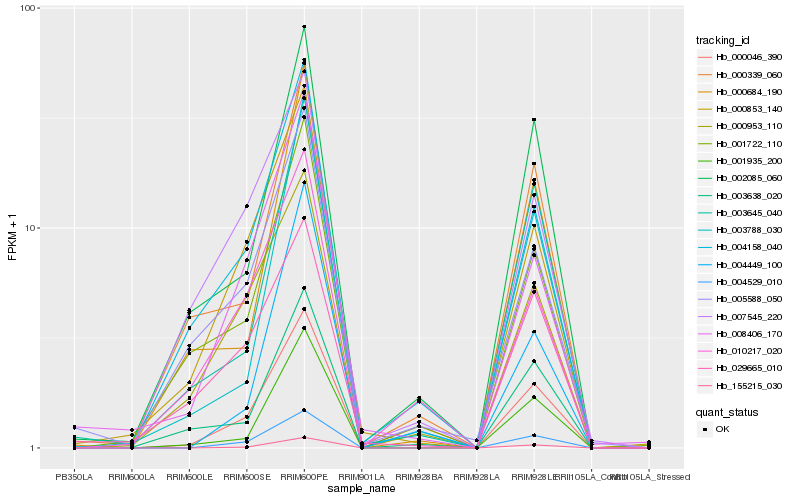
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004529_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_27055 [Jatropha curcas] |
| 2 |
Hb_155215_030 |
0.0705643188 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 3 |
Hb_000853_140 |
0.1258452894 |
- |
- |
hypothetical protein POPTR_0014s12080g [Populus trichocarpa] |
| 4 |
Hb_001935_200 |
0.1351752776 |
transcription factor |
TF Family: C2H2 |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_008406_170 |
0.1439767397 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_000953_110 |
0.1473762902 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105629453 [Jatropha curcas] |
| 7 |
Hb_004158_040 |
0.1546531517 |
- |
- |
PREDICTED: WAT1-related protein At2g37460-like [Jatropha curcas] |
| 8 |
Hb_002085_060 |
0.1610455385 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003645_040 |
0.1626034615 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0003s13190g [Populus trichocarpa] |
| 10 |
Hb_003638_020 |
0.1654342765 |
- |
- |
PREDICTED: uncharacterized protein LOC105645807 [Jatropha curcas] |
| 11 |
Hb_003788_030 |
0.1665704762 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_001722_110 |
0.1666678191 |
- |
- |
DUF26 domain-containing protein 1 precursor, putative [Ricinus communis] |
| 13 |
Hb_029665_010 |
0.1703547275 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 14 |
Hb_000339_060 |
0.1704804234 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 15 |
Hb_007545_220 |
0.1710623293 |
- |
- |
PREDICTED: uncharacterized protein LOC105632565 [Jatropha curcas] |
| 16 |
Hb_004449_100 |
0.1712909698 |
- |
- |
- |
| 17 |
Hb_000684_190 |
0.1718207219 |
- |
- |
beta-1,3-glucuronyltransferase, putative [Ricinus communis] |
| 18 |
Hb_005588_050 |
0.1724382761 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_010217_020 |
0.1734012444 |
- |
- |
cinnamate 4-hydroxylase [Leucaena leucocephala] |
| 20 |
Hb_000046_390 |
0.174182202 |
- |
- |
PREDICTED: CASP-like protein 1D1 [Jatropha curcas] |