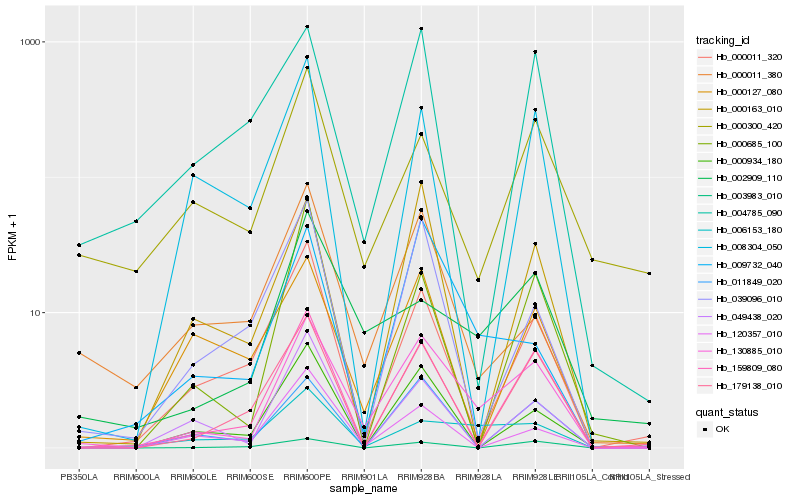
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_130885_010 |
0.0 |
- |
- |
PREDICTED: beta-glucosidase 13-like [Jatropha curcas] |
| 2 |
Hb_159809_080 |
0.1952812995 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 3 |
Hb_011849_020 |
0.2253043558 |
- |
- |
dioxygenase family protein [Populus trichocarpa] |
| 4 |
Hb_179138_010 |
0.2292592831 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 5 |
Hb_002909_110 |
0.2293528621 |
- |
- |
PREDICTED: uncharacterized protein LOC105648153 [Jatropha curcas] |
| 6 |
Hb_000011_320 |
0.229547138 |
- |
- |
PREDICTED: uncharacterized protein LOC105631328 [Jatropha curcas] |
| 7 |
Hb_000934_180 |
0.2298471319 |
- |
- |
hypothetical protein JCGZ_25237 [Jatropha curcas] |
| 8 |
Hb_009732_040 |
0.2304634253 |
- |
- |
hypothetical protein POPTR_0006s04880g [Populus trichocarpa] |
| 9 |
Hb_006153_180 |
0.231959544 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000163_010 |
0.2381770929 |
- |
- |
hypothetical protein JCGZ_13952 [Jatropha curcas] |
| 11 |
Hb_004785_090 |
0.2433615218 |
- |
- |
hypothetical protein MIMGU_mgv1a018797mg [Erythranthe guttata] |
| 12 |
Hb_039096_010 |
0.2439105027 |
- |
- |
unknown [Populus trichocarpa] |
| 13 |
Hb_000011_380 |
0.2465976467 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: subtilisin-like protease SBT1.6 [Jatropha curcas] |
| 14 |
Hb_008304_050 |
0.2468502958 |
- |
- |
PREDICTED: trans-cinnamate 4-monooxygenase [Jatropha curcas] |
| 15 |
Hb_000127_080 |
0.2480098738 |
- |
- |
hypothetical protein POPTR_0005s21760g [Populus trichocarpa] |
| 16 |
Hb_003983_010 |
0.2485079512 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 17 |
Hb_000300_420 |
0.2503153652 |
- |
- |
5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, putative [Ricinus communis] |
| 18 |
Hb_120357_010 |
0.25151283 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 19 |
Hb_049438_020 |
0.2519315768 |
- |
- |
PREDICTED: peroxidase 20 [Jatropha curcas] |
| 20 |
Hb_000685_100 |
0.2543878662 |
- |
- |
hypothetical protein POPTR_0001s10630g [Populus trichocarpa] |