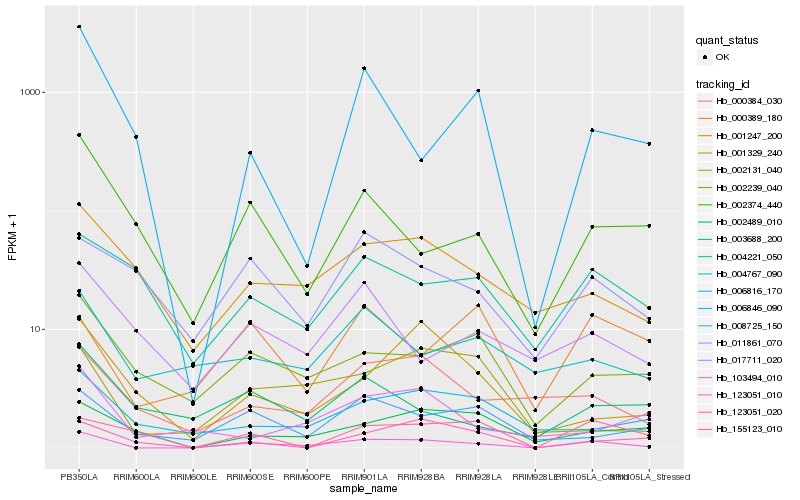
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_123051_010 |
0.0 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Populus euphratica] |
| 2 |
Hb_000384_030 |
0.2855617909 |
- |
- |
PREDICTED: uncharacterized protein LOC101310565 isoform X1 [Fragaria vesca subsp. vesca] |
| 3 |
Hb_001247_200 |
0.2897558263 |
- |
- |
PREDICTED: clathrin coat assembly protein AP180 [Jatropha curcas] |
| 4 |
Hb_002239_040 |
0.2931531979 |
- |
- |
Cellular apoptosis susceptibility protein / importin-alpha re-exporter, putative isoform 1 [Theobroma cacao] |
| 5 |
Hb_008725_150 |
0.3008724501 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 6 |
Hb_002131_040 |
0.303257734 |
- |
- |
proton-dependent oligopeptide transport family protein [Populus trichocarpa] |
| 7 |
Hb_002489_010 |
0.303979556 |
- |
- |
hypothetical protein JCGZ_17199 [Jatropha curcas] |
| 8 |
Hb_011861_070 |
0.3087614894 |
- |
- |
F-box/LRR-repeat protein, putative [Ricinus communis] |
| 9 |
Hb_002374_440 |
0.3097796945 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004767_090 |
0.3114609878 |
- |
- |
Aspartic proteinase Asp1 precursor, putative [Ricinus communis] |
| 11 |
Hb_006816_170 |
0.3125983214 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump 1-like [Jatropha curcas] |
| 12 |
Hb_123051_020 |
0.3138619707 |
- |
- |
hypothetical protein POPTR_2151s00200g [Populus trichocarpa] |
| 13 |
Hb_004221_050 |
0.3138952894 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g01540 [Jatropha curcas] |
| 14 |
Hb_001329_240 |
0.3152957276 |
- |
- |
hypothetical protein POPTR_0002s18920g [Populus trichocarpa] |
| 15 |
Hb_003688_200 |
0.3190924548 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_155123_010 |
0.319576314 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 17 |
Hb_006846_090 |
0.3206107104 |
- |
- |
PREDICTED: uncharacterized protein LOC105648695 [Jatropha curcas] |
| 18 |
Hb_017711_020 |
0.3217243394 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 56 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000389_180 |
0.3230794569 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 20 |
Hb_103494_010 |
0.3232869155 |
- |
- |
- |