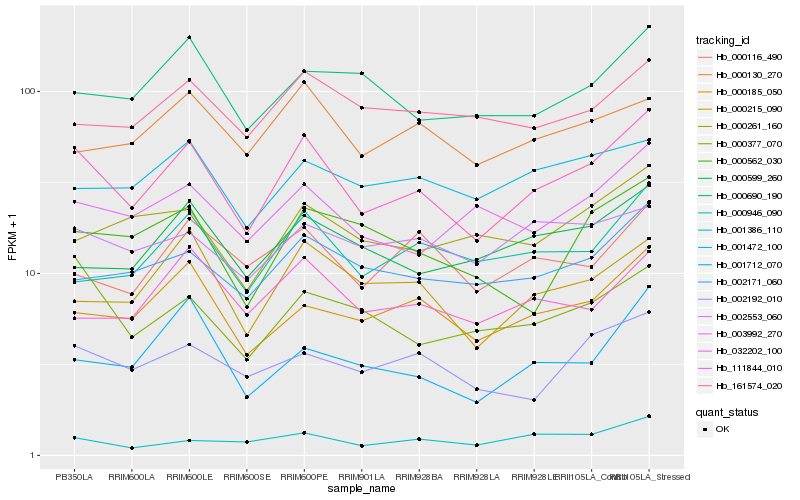
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_111844_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632109 [Jatropha curcas] |
| 2 |
Hb_000185_050 |
0.1165775865 |
- |
- |
PREDICTED: FKBP12-interacting protein of 37 kDa [Jatropha curcas] |
| 3 |
Hb_002171_060 |
0.1203198426 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000215_090 |
0.1208125551 |
- |
- |
cytochrome b5 domain-containing family protein [Populus trichocarpa] |
| 5 |
Hb_002192_010 |
0.1218663385 |
- |
- |
- |
| 6 |
Hb_003992_270 |
0.1221296674 |
- |
- |
PREDICTED: uncharacterized protein LOC105634861 isoform X3 [Jatropha curcas] |
| 7 |
Hb_000377_070 |
0.1231177495 |
- |
- |
PREDICTED: alpha-ketoglutarate-dependent dioxygenase alkB homolog 2 [Jatropha curcas] |
| 8 |
Hb_000946_090 |
0.1278188557 |
- |
- |
PREDICTED: cyclin-C1-2-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_000599_260 |
0.1301021138 |
- |
- |
Vacuolar protein sorting-associated protein 2 like 3 [Glycine soja] |
| 10 |
Hb_000690_190 |
0.1301255856 |
- |
- |
40S ribosomal protein S7, putative [Ricinus communis] |
| 11 |
Hb_032202_100 |
0.1321591959 |
desease resistance |
Gene Name: ArsA_ATPase |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 12 |
Hb_001386_110 |
0.1330936723 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: putative fasciclin-like arabinogalactan protein 20 [Populus euphratica] |
| 13 |
Hb_001472_100 |
0.1331982441 |
- |
- |
ubiquitin-conjugating enzyme h, putative [Ricinus communis] |
| 14 |
Hb_000562_030 |
0.1337576744 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000130_270 |
0.1340675023 |
- |
- |
PREDICTED: malate dehydrogenase, mitochondrial [Jatropha curcas] |
| 16 |
Hb_161574_020 |
0.1345312183 |
- |
- |
PREDICTED: ubiquitin fusion degradation protein 1 homolog [Jatropha curcas] |
| 17 |
Hb_000261_160 |
0.1347155749 |
- |
- |
PREDICTED: actin-related protein 2/3 complex subunit 4 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000116_490 |
0.1355450661 |
- |
- |
PREDICTED: WD repeat-containing protein 55 homolog [Jatropha curcas] |
| 19 |
Hb_001712_070 |
0.1363219846 |
- |
- |
PREDICTED: HD domain-containing protein 2 [Jatropha curcas] |
| 20 |
Hb_002553_060 |
0.1375566081 |
- |
- |
PREDICTED: electron transfer flavoprotein subunit beta, mitochondrial [Jatropha curcas] |