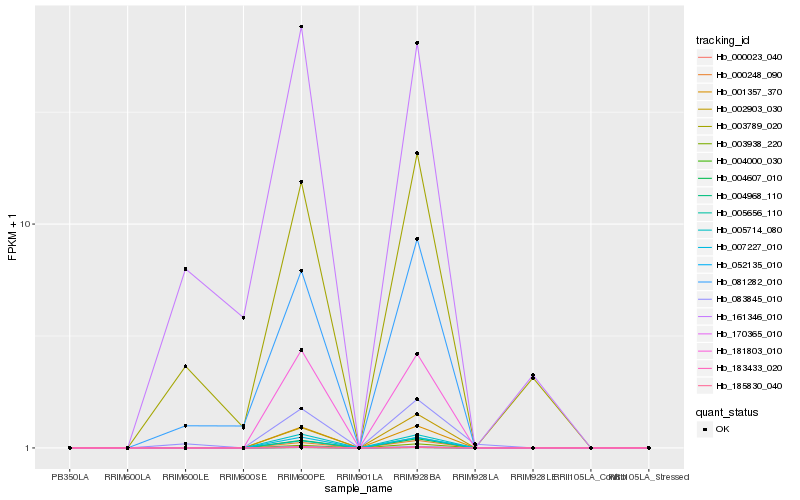
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_083845_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0015s09760g [Populus trichocarpa] |
| 2 |
Hb_081282_010 |
0.1666324489 |
- |
- |
PREDICTED: uncharacterized protein LOC105647834 [Jatropha curcas] |
| 3 |
Hb_003789_020 |
0.1829762181 |
- |
- |
hypothetical protein JCGZ_23602 [Jatropha curcas] |
| 4 |
Hb_004607_010 |
0.1855851424 |
- |
- |
- |
| 5 |
Hb_000248_090 |
0.1857450621 |
- |
- |
PREDICTED: uncharacterized protein LOC104110417 [Nicotiana tomentosiformis] |
| 6 |
Hb_005656_110 |
0.1868150714 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 7 |
Hb_003938_220 |
0.1892240124 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 8 |
Hb_001357_370 |
0.1899520025 |
- |
- |
- |
| 9 |
Hb_004968_110 |
0.191408944 |
- |
- |
hypothetical protein JCGZ_01361 [Jatropha curcas] |
| 10 |
Hb_000023_040 |
0.1918499473 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 11 |
Hb_161346_010 |
0.1925460152 |
- |
- |
PREDICTED: germin-like protein subfamily 1 member 20 [Populus euphratica] |
| 12 |
Hb_007227_010 |
0.194519892 |
- |
- |
hypothetical protein BN927_00890 [Lactococcus lactis subsp. lactis Dephy 1] |
| 13 |
Hb_005714_080 |
0.1958828317 |
transcription factor |
TF Family: B3 |
PREDICTED: uncharacterized protein LOC105109431 [Populus euphratica] |
| 14 |
Hb_002903_030 |
0.196389468 |
- |
- |
polyphenol oxidase [Cydonia oblonga] |
| 15 |
Hb_052135_010 |
0.196459844 |
- |
- |
PREDICTED: phytochrome B isoform X2 [Jatropha curcas] |
| 16 |
Hb_170365_010 |
0.1964927132 |
- |
- |
Retrotransposon protein, putative [Theobroma cacao] |
| 17 |
Hb_183433_020 |
0.1966914086 |
- |
- |
PREDICTED: uncharacterized protein LOC105851424 [Cicer arietinum] |
| 18 |
Hb_181803_010 |
0.1970951787 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 19 |
Hb_004000_030 |
0.1971200581 |
- |
- |
putative retrotransposon protein [Phyllostachys edulis] |
| 20 |
Hb_185830_040 |
0.1975162176 |
- |
- |
hypothetical protein JCGZ_16423 [Jatropha curcas] |