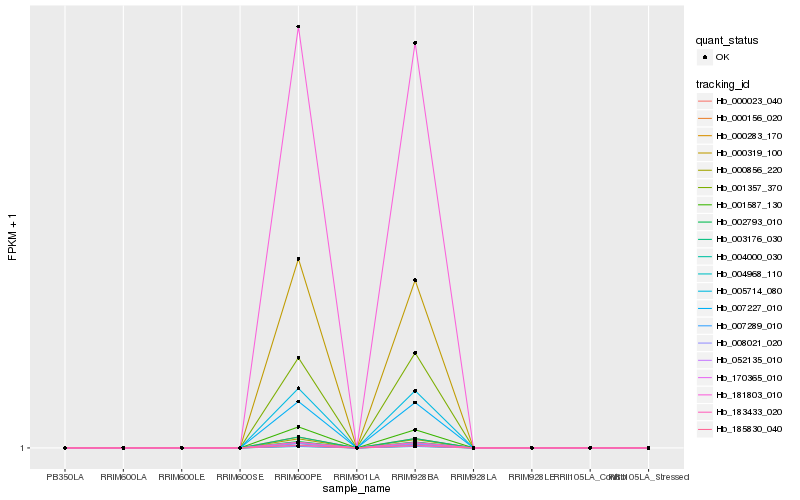
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007227_010 |
0.0 |
- |
- |
hypothetical protein BN927_00890 [Lactococcus lactis subsp. lactis Dephy 1] |
| 2 |
Hb_005714_080 |
0.0043779599 |
transcription factor |
TF Family: B3 |
PREDICTED: uncharacterized protein LOC105109431 [Populus euphratica] |
| 3 |
Hb_052135_010 |
0.006154257 |
- |
- |
PREDICTED: phytochrome B isoform X2 [Jatropha curcas] |
| 4 |
Hb_170365_010 |
0.0062541877 |
- |
- |
Retrotransposon protein, putative [Theobroma cacao] |
| 5 |
Hb_183433_020 |
0.0068554983 |
- |
- |
PREDICTED: uncharacterized protein LOC105851424 [Cicer arietinum] |
| 6 |
Hb_181803_010 |
0.0080631586 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 7 |
Hb_004000_030 |
0.0081369633 |
- |
- |
putative retrotransposon protein [Phyllostachys edulis] |
| 8 |
Hb_185830_040 |
0.0093029885 |
- |
- |
hypothetical protein JCGZ_16423 [Jatropha curcas] |
| 9 |
Hb_000023_040 |
0.0095365686 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 10 |
Hb_002793_010 |
0.009749751 |
- |
- |
putative metal-nicotianamine transporter YSL7 -like protein [Gossypium arboreum] |
| 11 |
Hb_000856_220 |
0.0101697536 |
- |
- |
hypothetical protein POPTR_0012s07300g [Populus trichocarpa] |
| 12 |
Hb_004968_110 |
0.0112730309 |
- |
- |
hypothetical protein JCGZ_01361 [Jatropha curcas] |
| 13 |
Hb_001357_370 |
0.0174667931 |
- |
- |
- |
| 14 |
Hb_007289_010 |
0.0174710607 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 15 |
Hb_008021_020 |
0.0176232246 |
transcription factor |
TF Family: GRF |
hypothetical protein POPTR_0014s00780g [Populus trichocarpa] |
| 16 |
Hb_000283_170 |
0.0179368801 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_000319_100 |
0.0263236105 |
- |
- |
PREDICTED: probable protein Pop3 [Jatropha curcas] |
| 18 |
Hb_001587_130 |
0.0266576726 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 19 |
Hb_000156_020 |
0.0277254497 |
- |
- |
PREDICTED: la-related protein 6A [Jatropha curcas] |
| 20 |
Hb_003176_030 |
0.0277254497 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Solanum lycopersicum] |