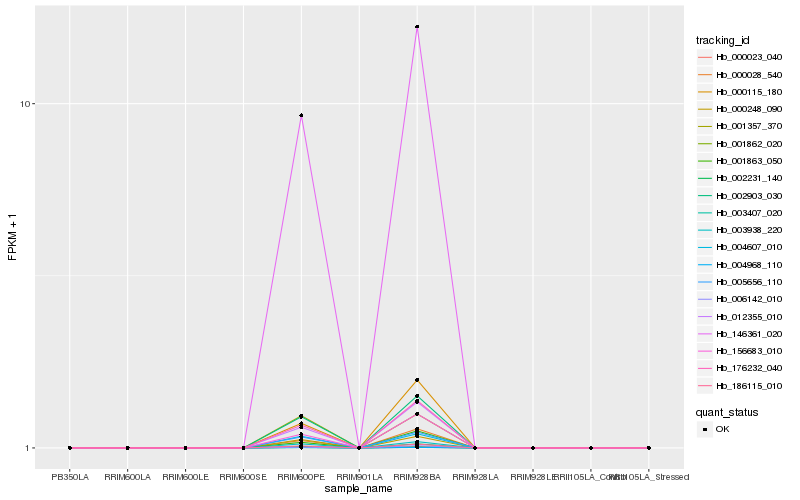
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_146361_020 |
0.0 |
- |
- |
PREDICTED: germin-like protein subfamily 1 member 11 [Jatropha curcas] |
| 2 |
Hb_186115_010 |
0.0016197955 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 3 |
Hb_002903_030 |
0.0121711649 |
- |
- |
polyphenol oxidase [Cydonia oblonga] |
| 4 |
Hb_001862_020 |
0.0130525914 |
- |
- |
PREDICTED: uncharacterized protein LOC103652604 [Zea mays] |
| 5 |
Hb_003407_020 |
0.0174413825 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 6 |
Hb_176232_040 |
0.0339146041 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003938_220 |
0.0389948428 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 8 |
Hb_012355_010 |
0.0472212003 |
- |
- |
PREDICTED: probable protein Pop3 [Jatropha curcas] |
| 9 |
Hb_005656_110 |
0.0532792259 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 10 |
Hb_000248_090 |
0.063246866 |
- |
- |
PREDICTED: uncharacterized protein LOC104110417 [Nicotiana tomentosiformis] |
| 11 |
Hb_156683_010 |
0.0648405613 |
- |
- |
PREDICTED: uncharacterized protein At5g65660-like [Jatropha curcas] |
| 12 |
Hb_000028_540 |
0.0668070371 |
- |
- |
hypothetical protein POPTR_0012s03720g [Populus trichocarpa] |
| 13 |
Hb_002231_140 |
0.0689363506 |
- |
- |
PREDICTED: uncharacterized protein LOC105634453 [Jatropha curcas] |
| 14 |
Hb_006142_010 |
0.0904917614 |
- |
- |
hypothetical protein CICLE_v10013815mg, partial [Citrus clementina] |
| 15 |
Hb_004607_010 |
0.0924757713 |
- |
- |
- |
| 16 |
Hb_001863_050 |
0.0967535241 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 8 [Jatropha curcas] |
| 17 |
Hb_000115_180 |
0.1001599031 |
- |
- |
hypothetical protein RCOM_1308310 [Ricinus communis] |
| 18 |
Hb_001357_370 |
0.122163975 |
- |
- |
- |
| 19 |
Hb_004968_110 |
0.1283235109 |
- |
- |
hypothetical protein JCGZ_01361 [Jatropha curcas] |
| 20 |
Hb_000023_040 |
0.1300497268 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |