| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
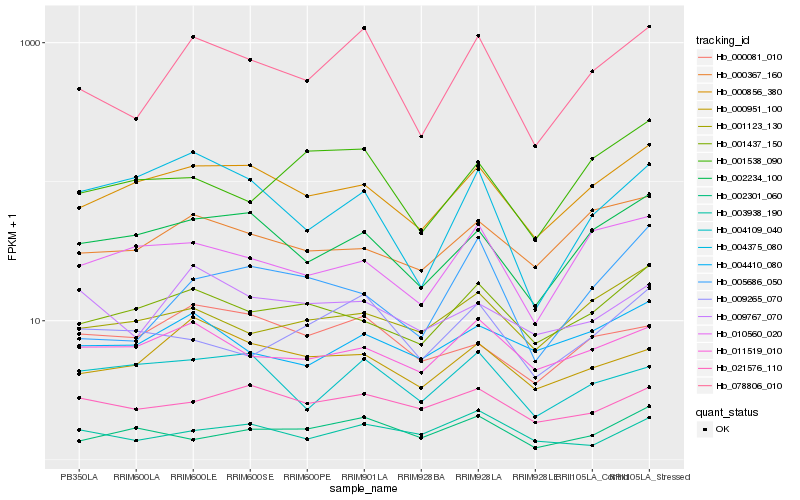
Hb_078806_010 |
0.0 |
- |
- |
hypothetical protein CISIN_1g031839mg [Citrus sinensis] |
| 2 |
Hb_000856_380 |
0.147248287 |
- |
- |
ubiquitin-conjugating enzyme [Medicago truncatula] |
| 3 |
Hb_002234_100 |
0.1534858434 |
- |
- |
F-box family protein, putative isoform 1 [Theobroma cacao] |
| 4 |
Hb_009265_070 |
0.1625002511 |
- |
- |
PREDICTED: uncharacterized protein LOC105649623 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000951_100 |
0.1633535435 |
- |
- |
PREDICTED: uncharacterized protein LOC103436565 isoform X1 [Malus domestica] |
| 6 |
Hb_001437_150 |
0.1697719161 |
- |
- |
PREDICTED: SNF1-related protein kinase regulatory subunit beta-3-like [Jatropha curcas] |
| 7 |
Hb_011519_010 |
0.1704009455 |
- |
- |
PREDICTED: uncharacterized protein LOC105645310 [Jatropha curcas] |
| 8 |
Hb_005686_050 |
0.1708172856 |
- |
- |
PREDICTED: probable tyrosine-protein phosphatase At1g05000 [Jatropha curcas] |
| 9 |
Hb_001538_090 |
0.1718852956 |
- |
- |
40S ribosomal protein S29 [Morus notabilis] |
| 10 |
Hb_010560_020 |
0.1726406771 |
- |
- |
PREDICTED: uncharacterized protein LOC105649267 [Jatropha curcas] |
| 11 |
Hb_000081_010 |
0.1739276254 |
- |
- |
hypothetical protein B456_012G056000 [Gossypium raimondii] |
| 12 |
Hb_009767_070 |
0.173983532 |
- |
- |
PREDICTED: uncharacterized protein LOC105631980 [Jatropha curcas] |
| 13 |
Hb_004410_080 |
0.1741257741 |
- |
- |
cellular nucleic acid binding protein, putative [Ricinus communis] |
| 14 |
Hb_000367_160 |
0.1741566264 |
- |
- |
PREDICTED: SNAP25 homologous protein SNAP33 [Populus euphratica] |
| 15 |
Hb_001123_130 |
0.1745918793 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 16 |
Hb_003938_190 |
0.1772175873 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g06145-like [Jatropha curcas] |
| 17 |
Hb_002301_060 |
0.179213217 |
- |
- |
hypothetical protein JCGZ_21483 [Jatropha curcas] |
| 18 |
Hb_004375_080 |
0.1793200896 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF073 [Jatropha curcas] |
| 19 |
Hb_021576_110 |
0.1799597007 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 4 [Jatropha curcas] |
| 20 |
Hb_004109_040 |
0.1805428881 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g53600, mitochondrial [Jatropha curcas] |