| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
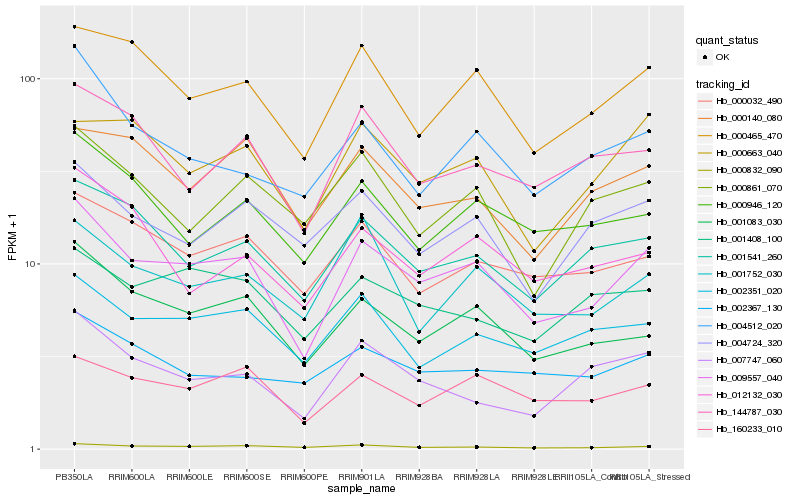
Hb_009557_040 |
0.0 |
- |
- |
PREDICTED: putative protein N-methyltransferase YNL024C [Jatropha curcas] |
| 2 |
Hb_001083_030 |
0.1001411259 |
- |
- |
PREDICTED: probable E3 ubiquitin ligase complex SCF subunit sconB [Jatropha curcas] |
| 3 |
Hb_160233_010 |
0.1017018365 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g08210 [Jatropha curcas] |
| 4 |
Hb_000465_470 |
0.1019679988 |
- |
- |
unknown [Picea sitchensis] |
| 5 |
Hb_000832_090 |
0.103723764 |
transcription factor |
TF Family: mTERF |
hypothetical protein VITISV_028994 [Vitis vinifera] |
| 6 |
Hb_001541_260 |
0.1115616958 |
- |
- |
PREDICTED: protein MAK16 homolog [Jatropha curcas] |
| 7 |
Hb_002351_020 |
0.1143853107 |
- |
- |
PREDICTED: rRNA methyltransferase 3B, mitochondrial [Jatropha curcas] |
| 8 |
Hb_000032_490 |
0.115176915 |
- |
- |
PREDICTED: uncharacterized protein At4g10930-like [Jatropha curcas] |
| 9 |
Hb_000946_120 |
0.1197564298 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 10 |
Hb_144787_030 |
0.1203760872 |
- |
- |
PREDICTED: protein SLOW GREEN 1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_004724_320 |
0.1230502484 |
- |
- |
Vesicle-associated membrane protein, putative [Ricinus communis] |
| 12 |
Hb_001408_100 |
0.1235510041 |
- |
- |
pseudouridylate synthase, putative [Ricinus communis] |
| 13 |
Hb_000663_040 |
0.1237872487 |
- |
- |
PREDICTED: uncharacterized protein LOC105650729 isoform X1 [Jatropha curcas] |
| 14 |
Hb_004512_020 |
0.1272216336 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 1 [Jatropha curcas] |
| 15 |
Hb_001752_030 |
0.128495034 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 4 [Jatropha curcas] |
| 16 |
Hb_007747_060 |
0.1286584215 |
- |
- |
PREDICTED: mitochondrial fission protein ELM1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002367_130 |
0.1296255027 |
- |
- |
UNC-50 family protein isoform 1 [Theobroma cacao] |
| 18 |
Hb_000140_080 |
0.1299780693 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_012132_030 |
0.1303988492 |
- |
- |
PREDICTED: RNA polymerase-associated protein CTR9 homolog [Jatropha curcas] |
| 20 |
Hb_000861_070 |
0.1308132327 |
- |
- |
conserved hypothetical protein [Ricinus communis] |