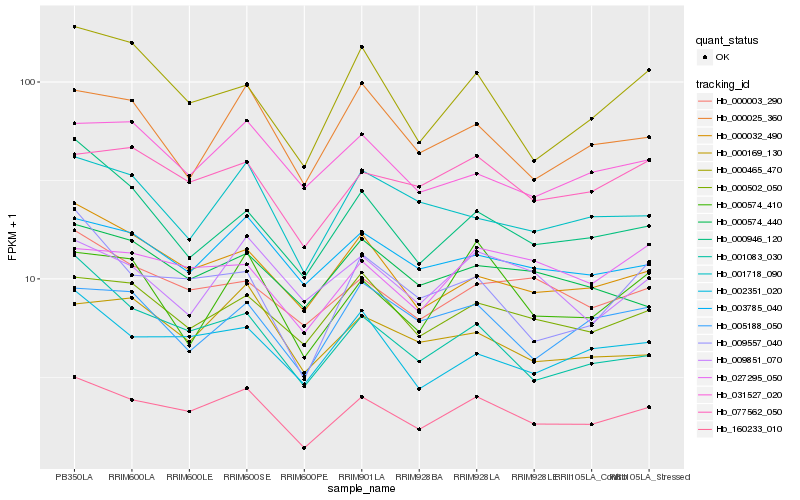
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_160233_010 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g08210 [Jatropha curcas] |
| 2 |
Hb_000032_490 |
0.0872313581 |
- |
- |
PREDICTED: uncharacterized protein At4g10930-like [Jatropha curcas] |
| 3 |
Hb_077562_050 |
0.0903129574 |
transcription factor |
TF Family: NF-YC |
PREDICTED: dr1-associated corepressor homolog [Jatropha curcas] |
| 4 |
Hb_000502_050 |
0.0909144768 |
transcription factor |
TF Family: HSF |
Heat shock factor protein HSF8, putative [Ricinus communis] |
| 5 |
Hb_003785_040 |
0.0916652575 |
- |
- |
PREDICTED: R3H domain-containing protein 2-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_002351_020 |
0.0924520845 |
- |
- |
PREDICTED: rRNA methyltransferase 3B, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000003_290 |
0.0973667479 |
- |
- |
hypothetical protein JCGZ_24486 [Jatropha curcas] |
| 8 |
Hb_009557_040 |
0.1017018365 |
- |
- |
PREDICTED: putative protein N-methyltransferase YNL024C [Jatropha curcas] |
| 9 |
Hb_000465_470 |
0.1017232438 |
- |
- |
unknown [Picea sitchensis] |
| 10 |
Hb_000025_360 |
0.1023281033 |
- |
- |
PREDICTED: protein SMG7 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001718_090 |
0.1034745282 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 97 [Jatropha curcas] |
| 12 |
Hb_031527_020 |
0.1041582468 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B isoform X1 [Jatropha curcas] |
| 13 |
Hb_000169_130 |
0.1046200427 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 14 |
Hb_027295_050 |
0.1051313413 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g48730, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000946_120 |
0.1051829309 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_005188_050 |
0.1054158254 |
- |
- |
PREDICTED: DNA-directed RNA polymerase 2B, chloroplastic/mitochondrial isoform X1 [Jatropha curcas] |
| 17 |
Hb_000574_410 |
0.1054778732 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g13600 [Jatropha curcas] |
| 18 |
Hb_001083_030 |
0.1062344009 |
- |
- |
PREDICTED: probable E3 ubiquitin ligase complex SCF subunit sconB [Jatropha curcas] |
| 19 |
Hb_000574_440 |
0.106475957 |
- |
- |
PREDICTED: uncharacterized protein LOC105634833 [Jatropha curcas] |
| 20 |
Hb_009851_070 |
0.1070802241 |
- |
- |
PREDICTED: cyclin-dependent kinase E-1 [Jatropha curcas] |