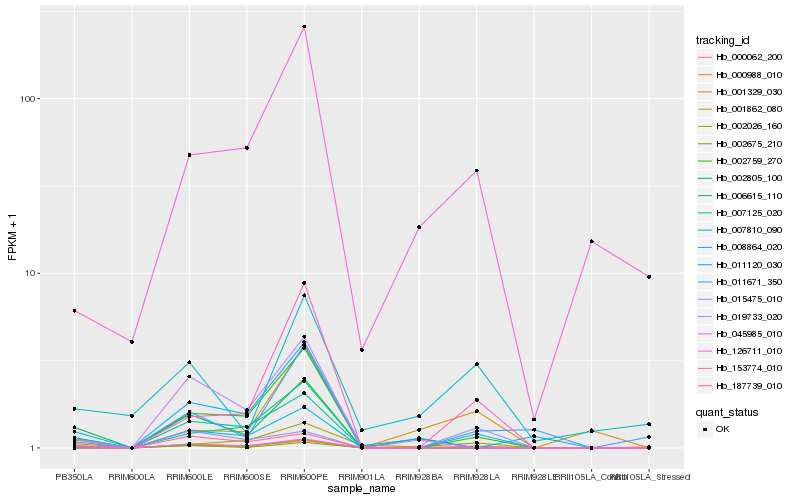
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008864_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_045985_010 |
0.2723863083 |
- |
- |
hypothetical protein JCGZ_00291 [Jatropha curcas] |
| 3 |
Hb_006615_110 |
0.2767319475 |
transcription factor |
TF Family: bZIP |
hypothetical protein JCGZ_11777 [Jatropha curcas] |
| 4 |
Hb_000988_010 |
0.2788961472 |
- |
- |
hypothetical protein POPTR_0004s17730g [Populus trichocarpa] |
| 5 |
Hb_002759_270 |
0.2825602883 |
- |
- |
Nectarin-1 precursor, putative [Ricinus communis] |
| 6 |
Hb_126711_010 |
0.3013723599 |
- |
- |
JAZ10 [Hevea brasiliensis] |
| 7 |
Hb_000062_200 |
0.3014265195 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 8 |
Hb_001862_080 |
0.3045112025 |
- |
- |
hypothetical protein RCOM_1154950 [Ricinus communis] |
| 9 |
Hb_187739_010 |
0.3111237264 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 10 |
Hb_002805_100 |
0.3203142566 |
- |
- |
Receptor-like protein 12 [Morus notabilis] |
| 11 |
Hb_015475_010 |
0.3232967216 |
- |
- |
hypothetical protein VITISV_034342 [Vitis vinifera] |
| 12 |
Hb_019733_020 |
0.3237933468 |
- |
- |
hypothetical protein POPTR_0003s15470g [Populus trichocarpa] |
| 13 |
Hb_002675_210 |
0.3256986051 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020-like [Citrus sinensis] |
| 14 |
Hb_002026_160 |
0.3271831616 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-like [Jatropha curcas] |
| 15 |
Hb_011671_350 |
0.3281379612 |
- |
- |
PREDICTED: uncharacterized protein LOC105648659 [Jatropha curcas] |
| 16 |
Hb_153774_010 |
0.3298954712 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_007125_020 |
0.3312678374 |
- |
- |
hypothetical protein POPTR_0003s18010g [Populus trichocarpa] |
| 18 |
Hb_001329_030 |
0.3314841708 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_011120_030 |
0.3325592524 |
- |
- |
PREDICTED: uncharacterized protein LOC105638239 [Jatropha curcas] |
| 20 |
Hb_007810_090 |
0.3327331541 |
- |
- |
actin actin-like protein [Coniophora puteana RWD-64-598 SS2] |