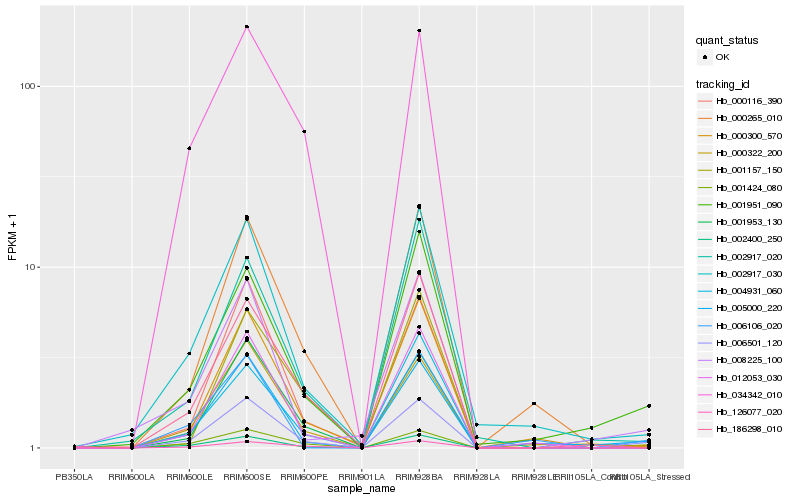
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006501_120 |
0.0 |
- |
- |
hypothetical protein JCGZ_07378 [Jatropha curcas] |
| 2 |
Hb_005000_220 |
0.1325017771 |
- |
- |
gulonolactone oxidase, putative [Ricinus communis] |
| 3 |
Hb_001951_090 |
0.1457763385 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 4 |
Hb_000322_200 |
0.1501161462 |
transcription factor |
TF Family: bHLH |
PREDICTED: putative transcription factor bHLH041 [Jatropha curcas] |
| 5 |
Hb_002400_250 |
0.1562361715 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 6 |
Hb_001953_130 |
0.1576365873 |
- |
- |
PREDICTED: uncharacterized protein LOC105648901 [Jatropha curcas] |
| 7 |
Hb_000116_390 |
0.1588773428 |
- |
- |
Mlo protein [Hevea brasiliensis] |
| 8 |
Hb_002917_030 |
0.1606202754 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase HSL2 [Setaria italica] |
| 9 |
Hb_000300_570 |
0.1648203389 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 10 |
Hb_012053_030 |
0.1686787524 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g22810 [Jatropha curcas] |
| 11 |
Hb_186298_010 |
0.1726537102 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_001424_080 |
0.1727964094 |
- |
- |
PREDICTED: probable L-type lectin-domain containing receptor kinase S.5 [Jatropha curcas] |
| 13 |
Hb_126077_020 |
0.1732181599 |
- |
- |
Kinase family protein with leucine-rich repeat domain [Theobroma cacao] |
| 14 |
Hb_000265_010 |
0.1743115993 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 15 |
Hb_008225_100 |
0.1748514989 |
- |
- |
- |
| 16 |
Hb_002917_020 |
0.1821144053 |
- |
- |
predicted protein [Arabidopsis lyrata subsp. lyrata] |
| 17 |
Hb_004931_060 |
0.1841481763 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-related protein Myb4 [Nelumbo nucifera] |
| 18 |
Hb_006106_020 |
0.1851670715 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 19 |
Hb_034342_010 |
0.1864206313 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001157_150 |
0.1880890391 |
- |
- |
PREDICTED: transmembrane protein 45A-like [Jatropha curcas] |