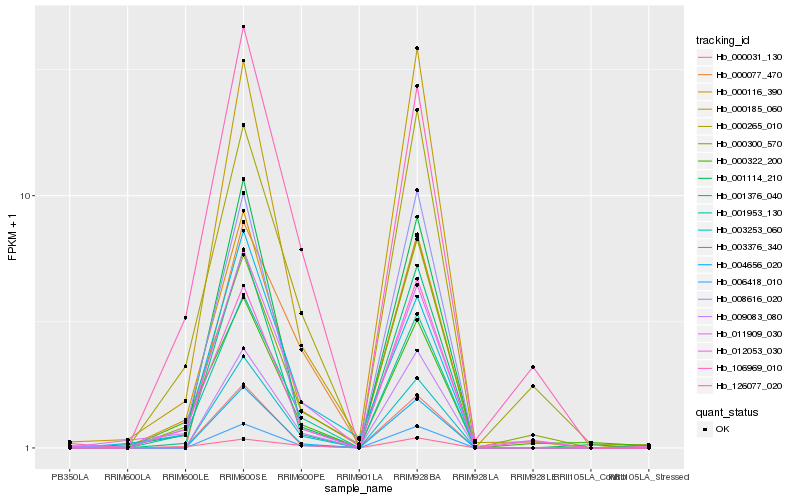
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001953_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648901 [Jatropha curcas] |
| 2 |
Hb_000116_390 |
0.0735571049 |
- |
- |
Mlo protein [Hevea brasiliensis] |
| 3 |
Hb_011909_030 |
0.1082464181 |
- |
- |
hypothetical protein JCGZ_00732 [Jatropha curcas] |
| 4 |
Hb_003253_060 |
0.1095048901 |
- |
- |
PREDICTED: tropinone reductase-like 1 [Vitis vinifera] |
| 5 |
Hb_000031_130 |
0.1096792457 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Populus euphratica] |
| 6 |
Hb_106969_010 |
0.1107712588 |
- |
- |
Cytochrome P450 76C4 [Glycine soja] |
| 7 |
Hb_012053_030 |
0.1109710458 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g22810 [Jatropha curcas] |
| 8 |
Hb_009083_080 |
0.1139295831 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000322_200 |
0.1175360025 |
transcription factor |
TF Family: bHLH |
PREDICTED: putative transcription factor bHLH041 [Jatropha curcas] |
| 10 |
Hb_001376_040 |
0.1199430117 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Populus euphratica] |
| 11 |
Hb_004656_020 |
0.1200858502 |
- |
- |
EF hand domain-containing protein, variant [Neurospora crassa OR74A] |
| 12 |
Hb_000265_010 |
0.1255971082 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 13 |
Hb_000185_060 |
0.1265249801 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001114_210 |
0.1296347385 |
- |
- |
PREDICTED: periaxin [Jatropha curcas] |
| 15 |
Hb_000300_570 |
0.1302070003 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 16 |
Hb_126077_020 |
0.131177265 |
- |
- |
Kinase family protein with leucine-rich repeat domain [Theobroma cacao] |
| 17 |
Hb_003376_340 |
0.1331298751 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000077_470 |
0.1332003188 |
- |
- |
PREDICTED: anthranilate N-methyltransferase-like [Jatropha curcas] |
| 19 |
Hb_008616_020 |
0.1336490948 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_006418_010 |
0.1369108901 |
- |
- |
PREDICTED: cytochrome P450 76A1-like [Jatropha curcas] |