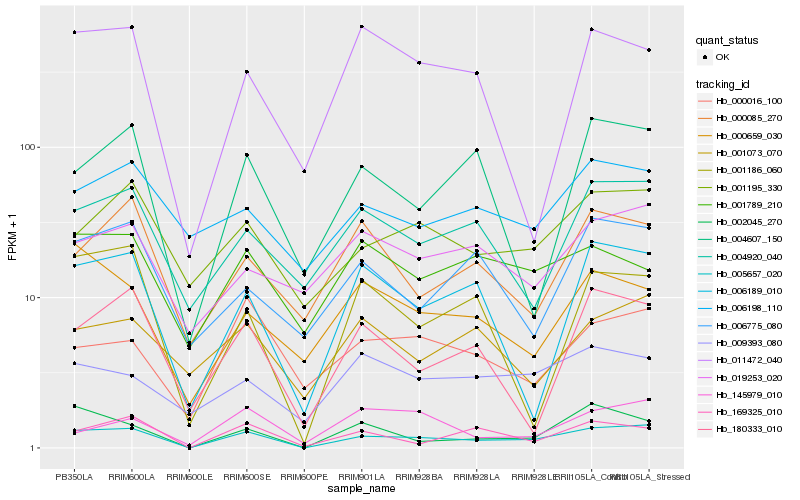
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005657_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_002045_270 |
0.1934897177 |
- |
- |
hypothetical protein JCGZ_21864 [Jatropha curcas] |
| 3 |
Hb_001195_330 |
0.1965408415 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_006189_010 |
0.1969857856 |
- |
- |
- |
| 5 |
Hb_169325_010 |
0.1994556621 |
- |
- |
PREDICTED: uncharacterized protein LOC103699787, partial [Phoenix dactylifera] |
| 6 |
Hb_004920_040 |
0.2039444406 |
- |
- |
PREDICTED: probable dimethyladenosine transferase [Jatropha curcas] |
| 7 |
Hb_180333_010 |
0.2092763215 |
- |
- |
- |
| 8 |
Hb_000016_100 |
0.2099546851 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP19 [Jatropha curcas] |
| 9 |
Hb_004607_150 |
0.2106061359 |
- |
- |
PREDICTED: serine/threonine-protein kinase UCNL [Jatropha curcas] |
| 10 |
Hb_001186_060 |
0.2116783223 |
- |
- |
- |
| 11 |
Hb_019253_020 |
0.2170913468 |
- |
- |
PREDICTED: uncharacterized protein LOC105647727 [Jatropha curcas] |
| 12 |
Hb_009393_080 |
0.2180731062 |
- |
- |
plant organelle RNA recognition domain protein [Medicago truncatula] |
| 13 |
Hb_145979_010 |
0.2186282583 |
- |
- |
PREDICTED: uncharacterized protein LOC105172316 [Sesamum indicum] |
| 14 |
Hb_000659_030 |
0.2191299289 |
- |
- |
pollen Ole e 1 allergen and extensin family protein, partial [Manihot esculenta] |
| 15 |
Hb_006775_080 |
0.219749935 |
- |
- |
PREDICTED: uncharacterized protein LOC105649730 [Jatropha curcas] |
| 16 |
Hb_001073_070 |
0.2198198409 |
- |
- |
PREDICTED: uncharacterized protein LOC105644546 isoform X1 [Jatropha curcas] |
| 17 |
Hb_006198_110 |
0.2200565052 |
- |
- |
PREDICTED: uncharacterized protein At4g13200, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000085_270 |
0.2213113459 |
- |
- |
PREDICTED: pirin-like protein [Malus domestica] |
| 19 |
Hb_001789_210 |
0.2225131202 |
- |
- |
PREDICTED: methyltransferase-like protein 1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_011472_040 |
0.2225877607 |
- |
- |
PREDICTED: clathrin light chain 3 [Jatropha curcas] |