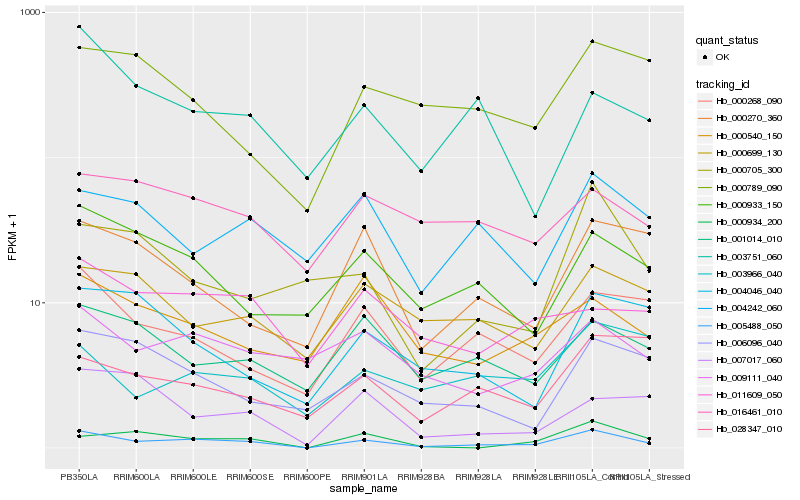
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005488_050 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: retrovirus-related Pol polyprotein from transposon TNT 1-94 [Cucumis sativus] |
| 2 |
Hb_000268_090 |
0.2120289206 |
- |
- |
PREDICTED: uncharacterized protein LOC105628378 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000933_150 |
0.2236365788 |
- |
- |
PREDICTED: mitotic checkpoint protein BUB3.3 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000934_200 |
0.2270474544 |
- |
- |
PREDICTED: uncharacterized protein LOC105633952 [Jatropha curcas] |
| 5 |
Hb_009111_040 |
0.2298947557 |
- |
- |
PREDICTED: protein downstream neighbor of Son [Jatropha curcas] |
| 6 |
Hb_001014_010 |
0.2300407298 |
- |
- |
PREDICTED: uncharacterized protein LOC105647311 [Jatropha curcas] |
| 7 |
Hb_000270_360 |
0.2307393154 |
- |
- |
PREDICTED: serine/threonine-protein kinase pakF-like [Jatropha curcas] |
| 8 |
Hb_006096_040 |
0.2324814833 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 8 [Jatropha curcas] |
| 9 |
Hb_004242_060 |
0.2378420002 |
- |
- |
PREDICTED: nicotinamidase 2-like [Jatropha curcas] |
| 10 |
Hb_000705_300 |
0.2391979891 |
- |
- |
PREDICTED: probable protein phosphatase 2C 24 [Jatropha curcas] |
| 11 |
Hb_000789_090 |
0.2397512853 |
- |
- |
hypothetical protein 17 [Hevea brasiliensis] |
| 12 |
Hb_004046_040 |
0.2402309486 |
transcription factor |
TF Family: GRF |
PREDICTED: growth-regulating factor 4-like [Jatropha curcas] |
| 13 |
Hb_000699_130 |
0.2404434509 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000540_150 |
0.2425349198 |
- |
- |
PREDICTED: uncharacterized protein YKR070W [Jatropha curcas] |
| 15 |
Hb_003751_060 |
0.2442558318 |
- |
- |
EARLY-RESPONSIVE TO DEHYDRATION 7 family protein [Populus trichocarpa] |
| 16 |
Hb_011609_050 |
0.2448560302 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_003966_040 |
0.2452028177 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2 [Jatropha curcas] |
| 18 |
Hb_007017_060 |
0.2460663555 |
- |
- |
PREDICTED: 5-formyltetrahydrofolate cyclo-ligase, mitochondrial-like [Jatropha curcas] |
| 19 |
Hb_016461_010 |
0.2471973385 |
- |
- |
PREDICTED: proline synthase co-transcribed bacterial homolog protein [Jatropha curcas] |
| 20 |
Hb_028347_010 |
0.2478259365 |
- |
- |
- |