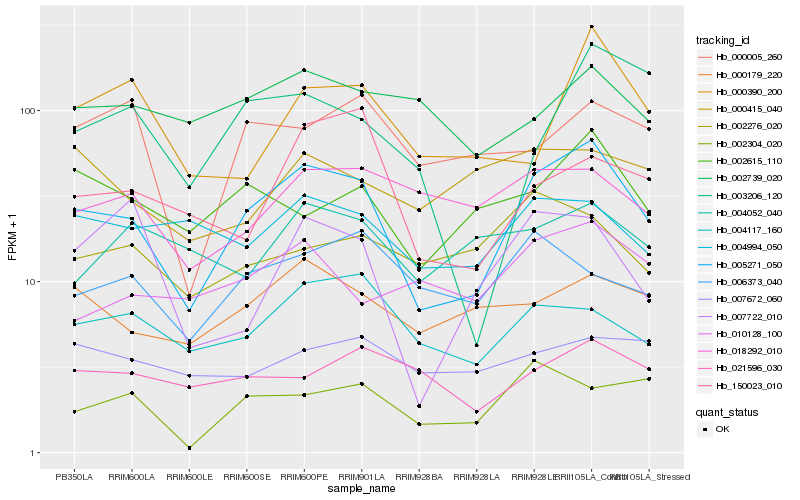
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005271_050 |
0.0 |
- |
- |
Tubulin beta-8 chain -like protein [Gossypium arboreum] |
| 2 |
Hb_004117_160 |
0.1902438321 |
transcription factor |
TF Family: B3 |
B3 domain-containing transcription repressor VAL2 -like protein [Gossypium arboreum] |
| 3 |
Hb_002615_110 |
0.1935830142 |
- |
- |
PREDICTED: uncharacterized protein LOC105628989 [Jatropha curcas] |
| 4 |
Hb_000179_220 |
0.19462314 |
- |
- |
PREDICTED: uncharacterized membrane protein C776.05 [Jatropha curcas] |
| 5 |
Hb_002304_020 |
0.1991616117 |
- |
- |
- |
| 6 |
Hb_004994_050 |
0.201449414 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 7 |
Hb_150023_010 |
0.2065915484 |
- |
- |
50S ribosomal protein L2, putative [Ricinus communis] |
| 8 |
Hb_018292_010 |
0.2072625586 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-3 [Jatropha curcas] |
| 9 |
Hb_006373_040 |
0.2101109507 |
- |
- |
PREDICTED: potassium transporter 11-like [Nicotiana tomentosiformis] |
| 10 |
Hb_000390_200 |
0.211684626 |
- |
- |
PREDICTED: uncharacterized protein LOC105632462 [Jatropha curcas] |
| 11 |
Hb_007722_010 |
0.2123975631 |
- |
- |
PREDICTED: kinesin light chain [Jatropha curcas] |
| 12 |
Hb_003206_120 |
0.2125604424 |
- |
- |
PREDICTED: glutamate decarboxylase 5-like [Jatropha curcas] |
| 13 |
Hb_010128_100 |
0.2131436903 |
- |
- |
PREDICTED: probable L-cysteine desulfhydrase, chloroplastic [Jatropha curcas] |
| 14 |
Hb_021596_030 |
0.2131645254 |
- |
- |
PREDICTED: uncharacterized protein LOC105631841 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002739_020 |
0.2140559058 |
- |
- |
PREDICTED: UTP--glucose-1-phosphate uridylyltransferase [Jatropha curcas] |
| 16 |
Hb_000005_260 |
0.2145128838 |
- |
- |
transferase, putative [Ricinus communis] |
| 17 |
Hb_002276_020 |
0.2161743494 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH49 isoform X1 [Jatropha curcas] |
| 18 |
Hb_007672_060 |
0.2199064788 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
Upstream activation factor subunit UAF30, putative [Ricinus communis] |
| 19 |
Hb_004052_040 |
0.2201103916 |
- |
- |
PREDICTED: putative G3BP-like protein [Jatropha curcas] |
| 20 |
Hb_000415_040 |
0.2203178398 |
- |
- |
PREDICTED: clathrin interactor EPSIN 2 [Jatropha curcas] |