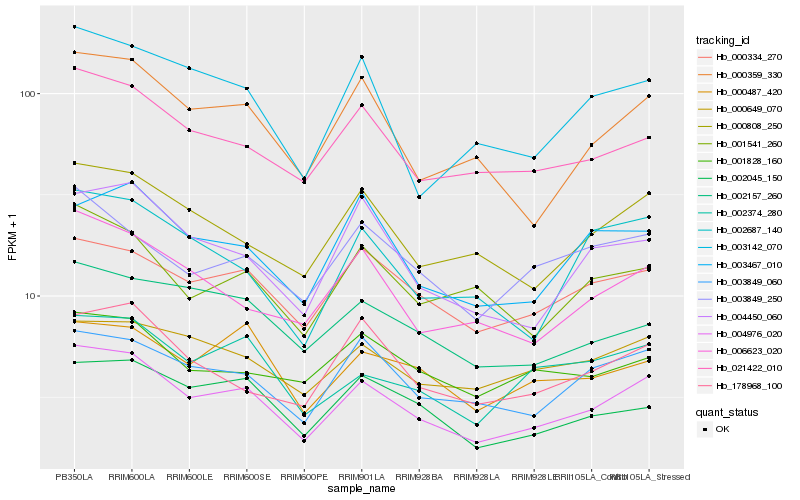
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004976_020 |
0.0 |
- |
- |
PREDICTED: probable nucleolar protein 5-1 [Jatropha curcas] |
| 2 |
Hb_021422_010 |
0.08199813 |
- |
- |
PREDICTED: alanine--glyoxylate aminotransferase 2 homolog 1, mitochondrial [Jatropha curcas] |
| 3 |
Hb_003849_060 |
0.082409267 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g13040, mitochondrial [Jatropha curcas] |
| 4 |
Hb_000808_250 |
0.082858012 |
- |
- |
PREDICTED: uncharacterized protein LOC105647419 [Jatropha curcas] |
| 5 |
Hb_000359_330 |
0.0833640179 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000334_270 |
0.0858949327 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_006623_020 |
0.0898100881 |
- |
- |
PREDICTED: protein LURP-one-related 7 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002045_150 |
0.091083941 |
- |
- |
histone acetyltransferase gcn5, putative [Ricinus communis] |
| 9 |
Hb_004450_060 |
0.0911342451 |
- |
- |
PREDICTED: uncharacterized protein LOC105638784 isoform X2 [Jatropha curcas] |
| 10 |
Hb_002374_280 |
0.0911680694 |
- |
- |
- |
| 11 |
Hb_000649_070 |
0.0957664016 |
- |
- |
PREDICTED: probable UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase 2 [Jatropha curcas] |
| 12 |
Hb_002157_260 |
0.0961565705 |
- |
- |
PREDICTED: partner of Y14 and mago-like [Jatropha curcas] |
| 13 |
Hb_001541_260 |
0.0963016931 |
- |
- |
PREDICTED: protein MAK16 homolog [Jatropha curcas] |
| 14 |
Hb_178968_100 |
0.0964487901 |
- |
- |
PREDICTED: putative glucose-6-phosphate 1-epimerase isoform X1 [Populus euphratica] |
| 15 |
Hb_003142_070 |
0.0971652159 |
- |
- |
hypothetical protein F383_14657 [Gossypium arboreum] |
| 16 |
Hb_000487_420 |
0.0980613039 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At1g04390 [Jatropha curcas] |
| 17 |
Hb_003467_010 |
0.1000014845 |
- |
- |
PREDICTED: probable thiol methyltransferase 2 [Jatropha curcas] |
| 18 |
Hb_003849_250 |
0.1001401618 |
- |
- |
PREDICTED: protein MAK16 homolog [Jatropha curcas] |
| 19 |
Hb_001828_160 |
0.1012617492 |
- |
- |
hypothetical protein F775_31105 [Aegilops tauschii] |
| 20 |
Hb_002687_140 |
0.1025978588 |
- |
- |
hypothetical protein JCGZ_24463 [Jatropha curcas] |