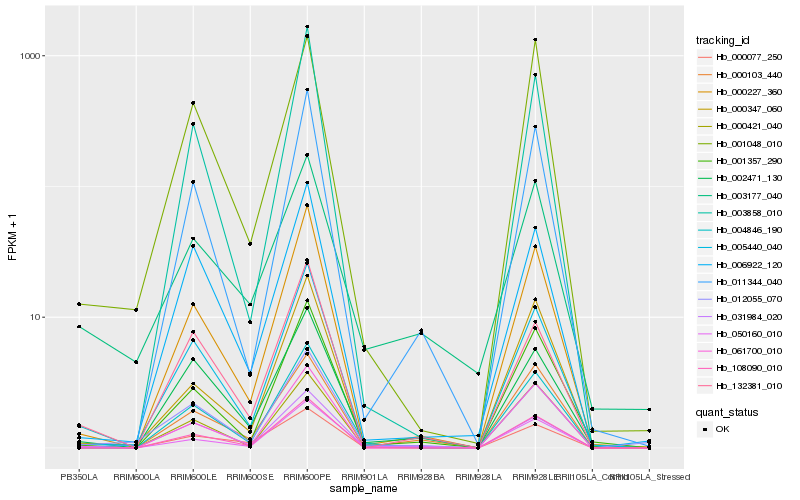
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004846_190 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s03430g [Populus trichocarpa] |
| 2 |
Hb_050160_010 |
0.129832235 |
- |
- |
hypothetical protein JCGZ_24934 [Jatropha curcas] |
| 3 |
Hb_031984_020 |
0.153001135 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000077_250 |
0.1533736301 |
- |
- |
hypothetical protein JCGZ_25802 [Jatropha curcas] |
| 5 |
Hb_000227_360 |
0.1561622018 |
- |
- |
PREDICTED: mavicyanin [Jatropha curcas] |
| 6 |
Hb_005440_040 |
0.1594164451 |
- |
- |
PREDICTED: microtubule-associated protein 70-5 [Jatropha curcas] |
| 7 |
Hb_132381_010 |
0.164042202 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 8 |
Hb_001357_290 |
0.169213766 |
- |
- |
PREDICTED: uncharacterized protein LOC105632986 [Jatropha curcas] |
| 9 |
Hb_108090_010 |
0.171599838 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002471_130 |
0.1716359446 |
- |
- |
PREDICTED: probable transmembrane ascorbate ferrireductase 3 [Jatropha curcas] |
| 11 |
Hb_012055_070 |
0.1731798224 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 12 |
Hb_003858_010 |
0.1748000958 |
- |
- |
xyloglucan endotransglucosylase/hydrolase [Gossypium hirsutum] |
| 13 |
Hb_003177_040 |
0.1758855449 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 14 |
Hb_061700_010 |
0.1780021342 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Prunus mume] |
| 15 |
Hb_000421_040 |
0.1786559805 |
- |
- |
Peptide transporter, putative [Ricinus communis] |
| 16 |
Hb_006922_120 |
0.1786652886 |
- |
- |
hypothetical protein JCGZ_01146 [Jatropha curcas] |
| 17 |
Hb_011344_040 |
0.1799575833 |
- |
- |
PREDICTED: 21 kDa protein [Jatropha curcas] |
| 18 |
Hb_000347_060 |
0.1808149506 |
- |
- |
PREDICTED: uncharacterized protein LOC105631356 [Jatropha curcas] |
| 19 |
Hb_000103_440 |
0.1813704019 |
- |
- |
hypothetical protein POPTR_0008s15900g [Populus trichocarpa] |
| 20 |
Hb_001048_010 |
0.1814690784 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 7 [Jatropha curcas] |