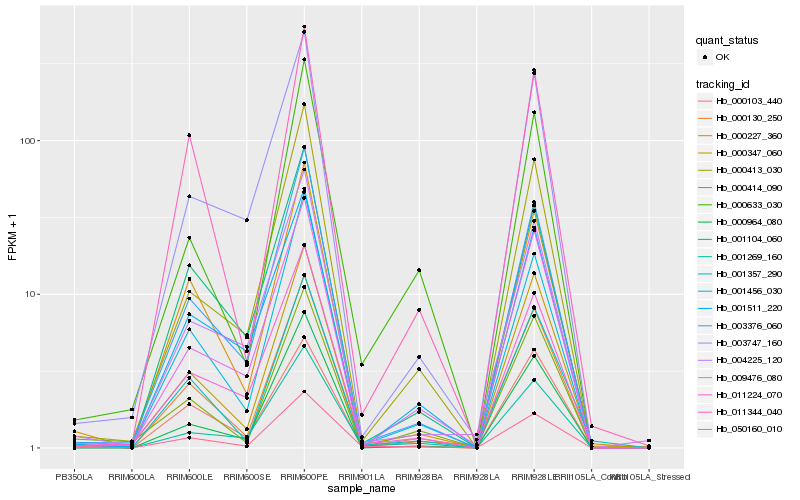
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_050160_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_24934 [Jatropha curcas] |
| 2 |
Hb_000227_360 |
0.0996455628 |
- |
- |
PREDICTED: mavicyanin [Jatropha curcas] |
| 3 |
Hb_001357_290 |
0.099655467 |
- |
- |
PREDICTED: uncharacterized protein LOC105632986 [Jatropha curcas] |
| 4 |
Hb_001456_030 |
0.1050800141 |
- |
- |
PREDICTED: bifunctional pinoresinol-lariciresinol reductase 2 [Vitis vinifera] |
| 5 |
Hb_003376_060 |
0.107648334 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000130_250 |
0.1090766006 |
- |
- |
PREDICTED: protein trichome birefringence-like 31 [Jatropha curcas] |
| 7 |
Hb_001269_160 |
0.1092579817 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000964_080 |
0.1096164941 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Jatropha curcas] |
| 9 |
Hb_000414_090 |
0.1099052913 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_011224_070 |
0.110345229 |
- |
- |
PREDICTED: solute carrier family 35 member E3 [Jatropha curcas] |
| 11 |
Hb_000633_030 |
0.1158791846 |
- |
- |
hypothetical protein POPTR_0003s21020g, partial [Populus trichocarpa] |
| 12 |
Hb_000347_060 |
0.118264722 |
- |
- |
PREDICTED: uncharacterized protein LOC105631356 [Jatropha curcas] |
| 13 |
Hb_000103_440 |
0.1189147122 |
- |
- |
hypothetical protein POPTR_0008s15900g [Populus trichocarpa] |
| 14 |
Hb_009476_080 |
0.1192274421 |
- |
- |
BnaA03g18920D [Brassica napus] |
| 15 |
Hb_000413_030 |
0.1222704695 |
- |
- |
PREDICTED: NAD(P)H dehydrogenase (quinone) FQR1-like [Jatropha curcas] |
| 16 |
Hb_011344_040 |
0.1226306285 |
- |
- |
PREDICTED: 21 kDa protein [Jatropha curcas] |
| 17 |
Hb_001104_060 |
0.1230848581 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9 [Jatropha curcas] |
| 18 |
Hb_004225_120 |
0.1233222341 |
- |
- |
PREDICTED: uncharacterized protein LOC105628059 [Jatropha curcas] |
| 19 |
Hb_001511_220 |
0.1253499693 |
- |
- |
hypothetical protein POPTR_0008s06990g [Populus trichocarpa] |
| 20 |
Hb_003747_160 |
0.1278505326 |
- |
- |
conserved hypothetical protein [Ricinus communis] |