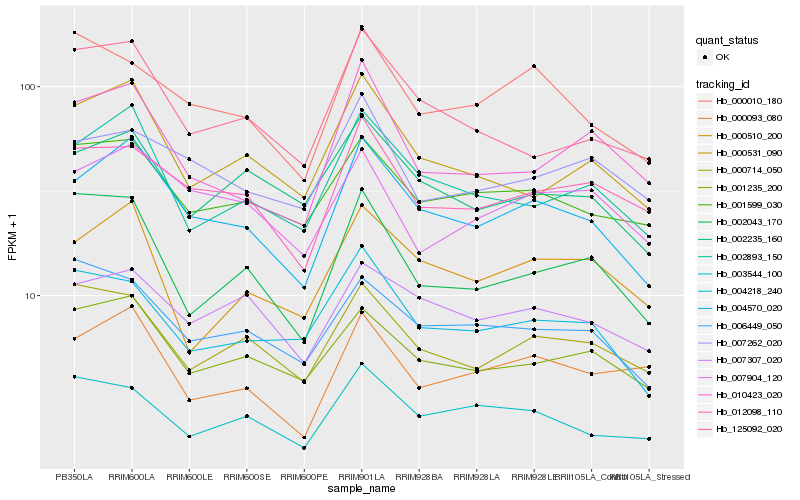
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004570_020 |
0.0 |
- |
- |
PREDICTED: protein TIC 100 [Jatropha curcas] |
| 2 |
Hb_002893_150 |
0.079931394 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 [Jatropha curcas] |
| 3 |
Hb_001235_200 |
0.093591751 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000531_090 |
0.0944841997 |
- |
- |
PREDICTED: transcription initiation factor IIF subunit alpha [Jatropha curcas] |
| 5 |
Hb_007307_020 |
0.0946339593 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_012098_110 |
0.094720081 |
- |
- |
Ribonuclease 3 [Gossypium arboreum] |
| 7 |
Hb_002235_160 |
0.0957082967 |
- |
- |
PREDICTED: SNF1-related protein kinase catalytic subunit alpha KIN10 [Jatropha curcas] |
| 8 |
Hb_001599_030 |
0.0971096773 |
- |
- |
PREDICTED: valine--tRNA ligase [Jatropha curcas] |
| 9 |
Hb_007904_120 |
0.1017474246 |
- |
- |
PREDICTED: ALBINO3-like protein 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_125092_020 |
0.1028552839 |
- |
- |
PREDICTED: nucleolin 1-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_010423_020 |
0.1044830966 |
- |
- |
PREDICTED: pre-mRNA-splicing factor CWC25 homolog [Jatropha curcas] |
| 12 |
Hb_000714_050 |
0.104803864 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g16880-like [Jatropha curcas] |
| 13 |
Hb_004218_240 |
0.1060665894 |
- |
- |
PREDICTED: DNA-directed RNA polymerase V subunit 1 [Jatropha curcas] |
| 14 |
Hb_000010_180 |
0.1068788954 |
- |
- |
PREDICTED: 28 kDa ribonucleoprotein, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_000510_200 |
0.1070726942 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 16 |
Hb_007262_020 |
0.1095855603 |
- |
- |
PREDICTED: protein AUXIN SIGNALING F-BOX 2-like [Jatropha curcas] |
| 17 |
Hb_000093_080 |
0.1096555018 |
- |
- |
hypothetical protein POPTR_0008s09730g [Populus trichocarpa] |
| 18 |
Hb_006449_050 |
0.1097100349 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ATX2-like [Jatropha curcas] |
| 19 |
Hb_002043_170 |
0.1102136414 |
- |
- |
PREDICTED: dentin sialophosphoprotein [Jatropha curcas] |
| 20 |
Hb_003544_100 |
0.1104828333 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g59900 [Jatropha curcas] |