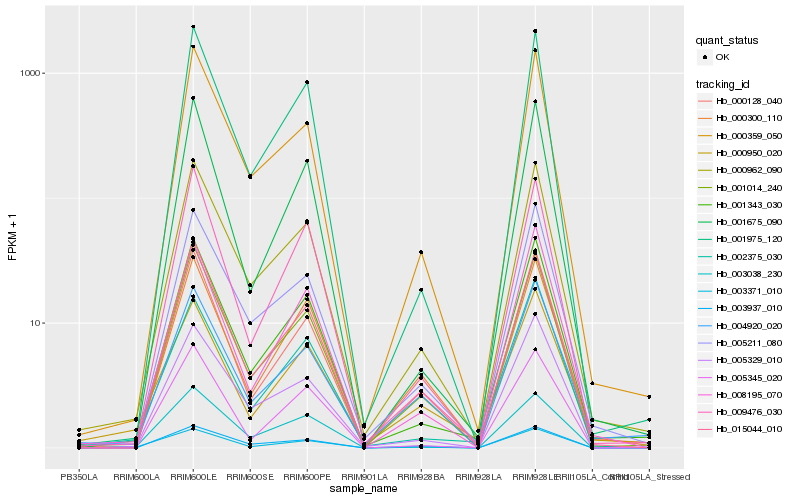
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003371_010 |
0.0 |
- |
- |
PREDICTED: putative cell division cycle ATPase [Jatropha curcas] |
| 2 |
Hb_000962_090 |
0.0960358649 |
- |
- |
omega-6 fatty acid desaturase [Hevea brasiliensis] |
| 3 |
Hb_001014_240 |
0.0961633241 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.1 [Jatropha curcas] |
| 4 |
Hb_000128_040 |
0.1030928647 |
- |
- |
PREDICTED: uncharacterized protein LOC105641228 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000950_020 |
0.1032429905 |
- |
- |
PREDICTED: glycerol-3-phosphate dehydrogenase [NAD(+)] GPDHC1, cytosolic [Jatropha curcas] |
| 6 |
Hb_015044_010 |
0.1053832926 |
- |
- |
hypothetical protein JCGZ_25130 [Jatropha curcas] |
| 7 |
Hb_003038_230 |
0.1072296018 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 7 [Jatropha curcas] |
| 8 |
Hb_001343_030 |
0.108407598 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit N, chloroplastic [Jatropha curcas] |
| 9 |
Hb_008195_070 |
0.1094754577 |
- |
- |
PREDICTED: thylakoid lumenal protein At1g12250, chloroplastic isoform X2 [Jatropha curcas] |
| 10 |
Hb_000359_050 |
0.1105784649 |
- |
- |
Oxygen-evolving enhancer protein 2, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_001975_120 |
0.1106245276 |
- |
- |
chlorophyll a/b binding protein type II [Glycine max] |
| 12 |
Hb_002375_030 |
0.1123037107 |
- |
- |
F-box family protein [Populus trichocarpa] |
| 13 |
Hb_000300_110 |
0.1129404728 |
- |
- |
hypothetical protein JCGZ_02651 [Jatropha curcas] |
| 14 |
Hb_005329_010 |
0.1147164898 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11410 [Jatropha curcas] |
| 15 |
Hb_005211_080 |
0.1148926042 |
- |
- |
hypothetical protein POPTR_0010s13740g [Populus trichocarpa] |
| 16 |
Hb_009476_030 |
0.1174833542 |
- |
- |
fatty acid desaturase [Manihot esculenta] |
| 17 |
Hb_005345_020 |
0.1178369881 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 18 |
Hb_001675_090 |
0.1188459213 |
- |
- |
Photosystem I reaction center subunit VI, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_004920_020 |
0.1204396378 |
- |
- |
hypothetical protein POPTR_0014s11700g [Populus trichocarpa] |
| 20 |
Hb_003937_010 |
0.1209031376 |
- |
- |
hypothetical protein JCGZ_20930 [Jatropha curcas] |