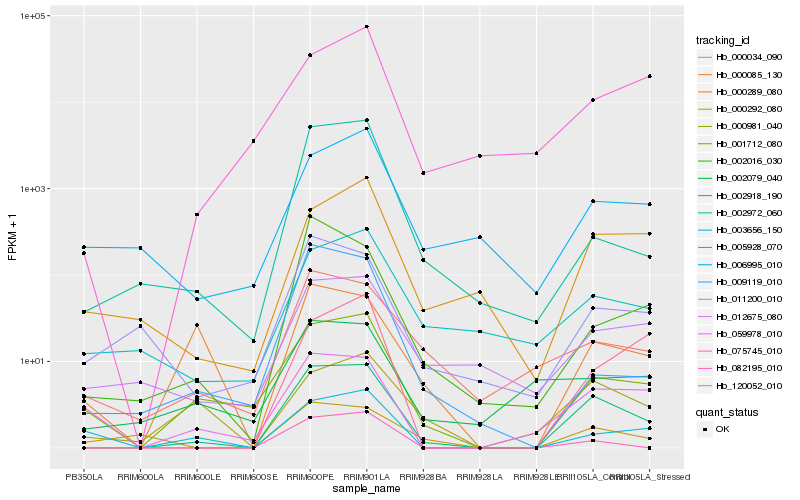
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002918_190 |
0.0 |
- |
- |
Uncharacterized protein TCM_029816 [Theobroma cacao] |
| 2 |
Hb_001712_080 |
0.2055917936 |
- |
- |
- |
| 3 |
Hb_082195_010 |
0.2210503126 |
- |
- |
Uncharacterized protein TCM_043808 [Theobroma cacao] |
| 4 |
Hb_002972_060 |
0.2425451043 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 5 |
Hb_000289_080 |
0.2458996593 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_011200_010 |
0.2464675519 |
- |
- |
- |
| 7 |
Hb_059978_010 |
0.2490041898 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X3 [Jatropha curcas] |
| 8 |
Hb_000034_090 |
0.2506233011 |
transcription factor |
TF Family: M-type |
MPF2-like, partial [Dunalia fasciculata] |
| 9 |
Hb_012675_080 |
0.2508821669 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 1 [Populus euphratica] |
| 10 |
Hb_120052_010 |
0.2535085052 |
- |
- |
hypothetical protein PRUPE_ppa1027171mg [Prunus persica] |
| 11 |
Hb_002016_030 |
0.2552085524 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000292_080 |
0.2569103316 |
- |
- |
hypothetical protein CISIN_1g034193mg [Citrus sinensis] |
| 13 |
Hb_003656_150 |
0.2609554585 |
- |
- |
- |
| 14 |
Hb_006995_010 |
0.2633828335 |
- |
- |
PREDICTED: 14 kDa zinc-binding protein [Jatropha curcas] |
| 15 |
Hb_000981_040 |
0.2648113909 |
- |
- |
- |
| 16 |
Hb_005928_070 |
0.2651812097 |
- |
- |
hypothetical protein POPTR_0002s04520g [Populus trichocarpa] |
| 17 |
Hb_000085_130 |
0.2658234768 |
- |
- |
- |
| 18 |
Hb_075745_010 |
0.2661695334 |
- |
- |
hypothetical protein EUGRSUZ_D00579 [Eucalyptus grandis] |
| 19 |
Hb_009119_010 |
0.2677089329 |
- |
- |
- |
| 20 |
Hb_002079_040 |
0.2731578856 |
- |
- |
hypothetical protein VITISV_018542 [Vitis vinifera] |