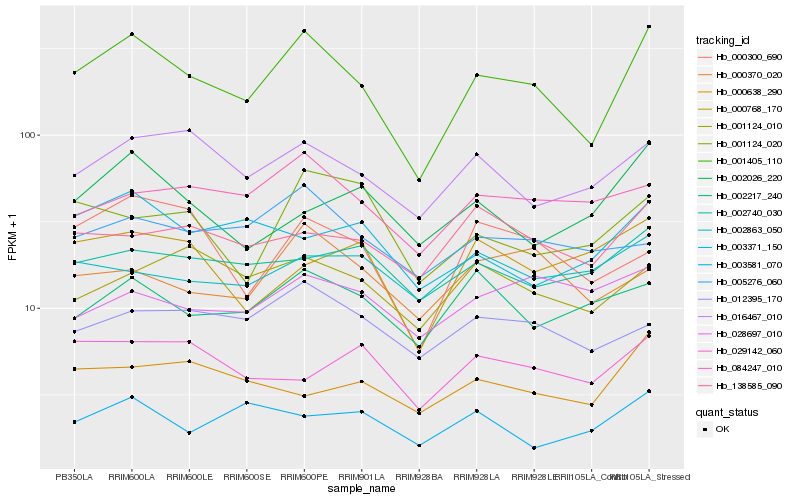
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001405_110 |
0.0 |
- |
- |
glutamine synthetase [Hevea brasiliensis] |
| 2 |
Hb_016467_010 |
0.1395362927 |
- |
- |
hypothetical protein POPTR_0004s18340g [Populus trichocarpa] |
| 3 |
Hb_084247_010 |
0.1526174674 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_012395_170 |
0.1527507892 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002217_240 |
0.1534267948 |
- |
- |
PREDICTED: F-box protein At5g49610 [Jatropha curcas] |
| 6 |
Hb_000300_690 |
0.15369573 |
- |
- |
hypothetical protein JCGZ_02719 [Jatropha curcas] |
| 7 |
Hb_138585_090 |
0.1537042063 |
- |
- |
Long chain base2 isoform 2 [Theobroma cacao] |
| 8 |
Hb_001124_010 |
0.1562012455 |
- |
- |
PREDICTED: serine/threonine-protein kinase WNK8 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000370_020 |
0.1570878541 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002740_030 |
0.1570970712 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002026_220 |
0.1604986246 |
- |
- |
Glutamate--cysteine ligase, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_005276_060 |
0.1607105156 |
- |
- |
PREDICTED: cyclin-P3-1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001124_020 |
0.1616136433 |
- |
- |
PREDICTED: protein yippee-like At4g27745 [Jatropha curcas] |
| 14 |
Hb_000638_290 |
0.1619399359 |
- |
- |
PREDICTED: 5' exonuclease Apollo [Jatropha curcas] |
| 15 |
Hb_003371_150 |
0.1620872846 |
- |
- |
PREDICTED: serine/threonine-protein kinase 10 [Jatropha curcas] |
| 16 |
Hb_000768_170 |
0.1672567878 |
- |
- |
hypothetical protein JCGZ_02624 [Jatropha curcas] |
| 17 |
Hb_028697_010 |
0.1673364907 |
- |
- |
PREDICTED: protein yippee-like At4g27745 [Jatropha curcas] |
| 18 |
Hb_029142_060 |
0.1678584367 |
- |
- |
PREDICTED: RNA pseudouridine synthase 2, chloroplastic [Jatropha curcas] |
| 19 |
Hb_003581_070 |
0.1685790296 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g40405 [Malus domestica] |
| 20 |
Hb_002863_050 |
0.1688539877 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |