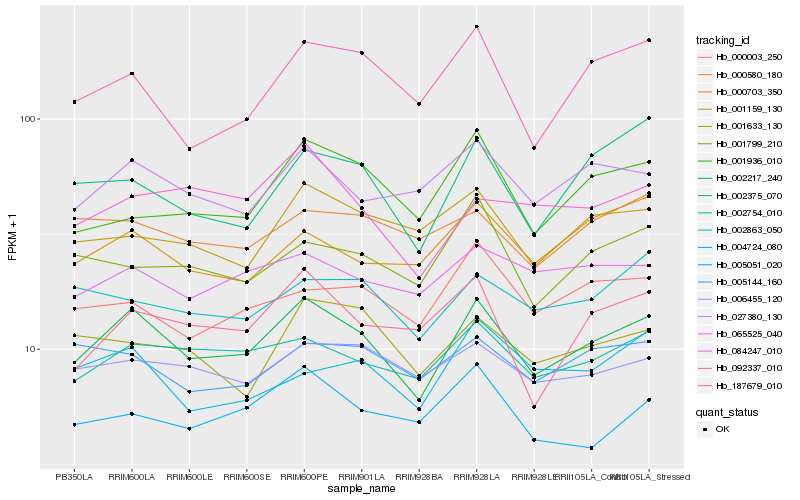
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002217_240 |
0.0 |
- |
- |
PREDICTED: F-box protein At5g49610 [Jatropha curcas] |
| 2 |
Hb_002754_010 |
0.0817264695 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670 [Jatropha curcas] |
| 3 |
Hb_027380_130 |
0.0874331983 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 23 isoform X2 [Jatropha curcas] |
| 4 |
Hb_006455_120 |
0.0959161638 |
- |
- |
n6-DNA-methyltransferase, putative [Ricinus communis] |
| 5 |
Hb_065525_040 |
0.0959209155 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3-like [Jatropha curcas] |
| 6 |
Hb_187679_010 |
0.0968601261 |
- |
- |
- |
| 7 |
Hb_001159_130 |
0.0976553462 |
- |
- |
PREDICTED: protein cornichon homolog 4 [Jatropha curcas] |
| 8 |
Hb_092337_010 |
0.0977078276 |
- |
- |
PREDICTED: actin-depolymerizing factor 2-like [Nelumbo nucifera] |
| 9 |
Hb_002863_050 |
0.0983285566 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 10 |
Hb_005051_020 |
0.0984293682 |
- |
- |
hypothetical protein JCGZ_20345 [Jatropha curcas] |
| 11 |
Hb_001799_210 |
0.0987330344 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 7 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002375_070 |
0.0994021317 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000003_250 |
0.0998480396 |
- |
- |
glycerol-3-phosphate dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_005144_160 |
0.1003560236 |
- |
- |
PREDICTED: uncharacterized protein LOC105628018 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001633_130 |
0.1006864884 |
- |
- |
PREDICTED: uncharacterized protein LOC105630200 [Jatropha curcas] |
| 16 |
Hb_084247_010 |
0.101275215 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000703_350 |
0.1013249768 |
- |
- |
PREDICTED: WD repeat-containing protein 26-like [Jatropha curcas] |
| 18 |
Hb_001936_010 |
0.1014200436 |
- |
- |
hypothetical protein Csa_2G348280 [Cucumis sativus] |
| 19 |
Hb_000580_180 |
0.1016622993 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004724_080 |
0.104617422 |
- |
- |
conserved hypothetical protein [Ricinus communis] |