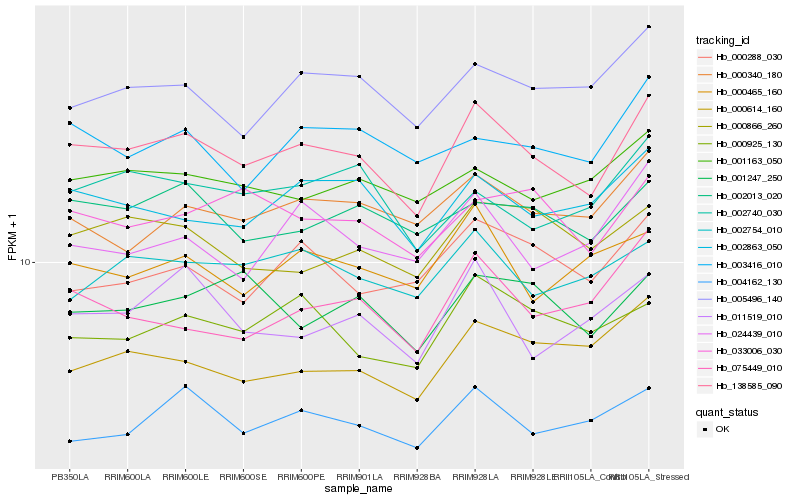
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_138585_090 |
0.0 |
- |
- |
Long chain base2 isoform 2 [Theobroma cacao] |
| 2 |
Hb_002863_050 |
0.0771385166 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 3 |
Hb_005496_140 |
0.0804690928 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000340_180 |
0.0822920946 |
- |
- |
PREDICTED: mitochondrial ubiquitin ligase activator of NFKB 1 [Jatropha curcas] |
| 5 |
Hb_002013_020 |
0.0824103852 |
- |
- |
PREDICTED: uncharacterized protein LOC105641613 [Jatropha curcas] |
| 6 |
Hb_000925_130 |
0.0840402916 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_004162_130 |
0.0840880071 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002754_010 |
0.0843000121 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670 [Jatropha curcas] |
| 9 |
Hb_033006_030 |
0.0843425449 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g22040-like [Jatropha curcas] |
| 10 |
Hb_001163_050 |
0.0866331931 |
- |
- |
PREDICTED: histone deacetylase 9 [Jatropha curcas] |
| 11 |
Hb_000866_260 |
0.0866743255 |
- |
- |
PREDICTED: F-box/kelch-repeat protein SKIP4 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000614_160 |
0.0870253394 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g04130, mitochondrial [Jatropha curcas] |
| 13 |
Hb_003416_010 |
0.0879182147 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 13 homolog B [Malus domestica] |
| 14 |
Hb_000288_030 |
0.088182987 |
- |
- |
PREDICTED: syntaxin-32 [Jatropha curcas] |
| 15 |
Hb_002740_030 |
0.0882989093 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_075449_010 |
0.0885977087 |
- |
- |
PREDICTED: cysteine synthase 2-like [Jatropha curcas] |
| 17 |
Hb_011519_010 |
0.0886016294 |
- |
- |
PREDICTED: uncharacterized protein LOC105645310 [Jatropha curcas] |
| 18 |
Hb_000465_160 |
0.0886263514 |
- |
- |
PREDICTED: uncharacterized protein LOC105632297 [Jatropha curcas] |
| 19 |
Hb_024439_010 |
0.089673676 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 2 [Jatropha curcas] |
| 20 |
Hb_001247_250 |
0.0906507026 |
- |
- |
PREDICTED: uncharacterized protein LOC105647377 [Jatropha curcas] |