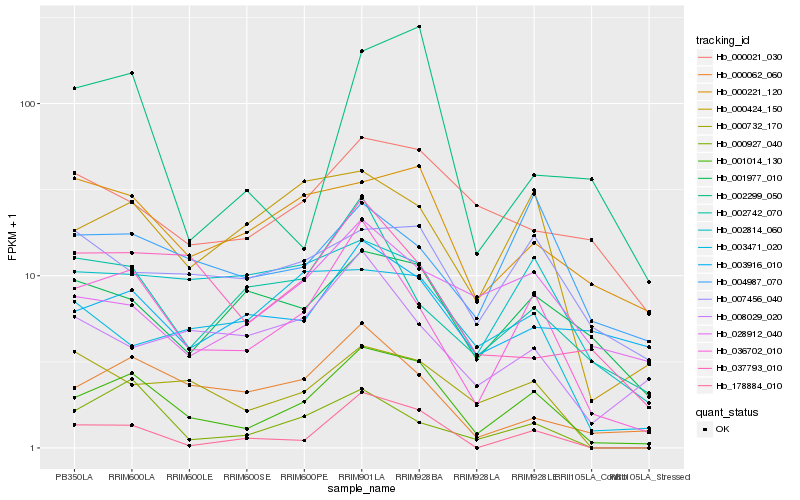
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001014_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647301 [Jatropha curcas] |
| 2 |
Hb_036702_010 |
0.1474716669 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 3 |
Hb_000424_150 |
0.2052329624 |
- |
- |
PREDICTED: copper transporter 5.1-like [Jatropha curcas] |
| 4 |
Hb_000062_060 |
0.2069801229 |
transcription factor |
TF Family: Orphans |
multi-sensor hybrid histidine kinase [Plautia stali symbiont] |
| 5 |
Hb_178884_010 |
0.2079514619 |
- |
- |
putative disease resistance gene NBS-LRR family protein [Populus trichocarpa] |
| 6 |
Hb_000732_170 |
0.2130801837 |
- |
- |
PREDICTED: putative PAP-specific phosphatase, mitochondrial isoform X1 [Jatropha curcas] |
| 7 |
Hb_000221_120 |
0.2191858565 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000927_040 |
0.2234372579 |
- |
- |
- |
| 9 |
Hb_001977_010 |
0.2243347919 |
- |
- |
PREDICTED: uncharacterized protein LOC105636485 isoform X2 [Jatropha curcas] |
| 10 |
Hb_002814_060 |
0.2263385333 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 [Jatropha curcas] |
| 11 |
Hb_000021_030 |
0.2305574482 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 12 |
Hb_007456_040 |
0.2313938455 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3B [Jatropha curcas] |
| 13 |
Hb_037793_010 |
0.2330702677 |
- |
- |
PREDICTED: microtubule-associated protein 70-2-like [Jatropha curcas] |
| 14 |
Hb_028912_040 |
0.2330976807 |
- |
- |
hypothetical protein POPTR_0016s03250g [Populus trichocarpa] |
| 15 |
Hb_003916_010 |
0.2332935768 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 13 [Jatropha curcas] |
| 16 |
Hb_004987_070 |
0.2337625039 |
- |
- |
PREDICTED: heat shock 70 kDa protein 17-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_002742_070 |
0.2370198593 |
- |
- |
PREDICTED: villin-4-like [Jatropha curcas] |
| 18 |
Hb_002299_050 |
0.2371297412 |
- |
- |
PREDICTED: chalcone--flavonone isomerase isoform X1 [Pyrus x bretschneideri] |
| 19 |
Hb_008029_020 |
0.2438132929 |
- |
- |
PREDICTED: uncharacterized protein LOC105631777 [Jatropha curcas] |
| 20 |
Hb_003471_020 |
0.2448660762 |
- |
- |
PREDICTED: ADP-ribosylation factor-like protein 8A [Jatropha curcas] |