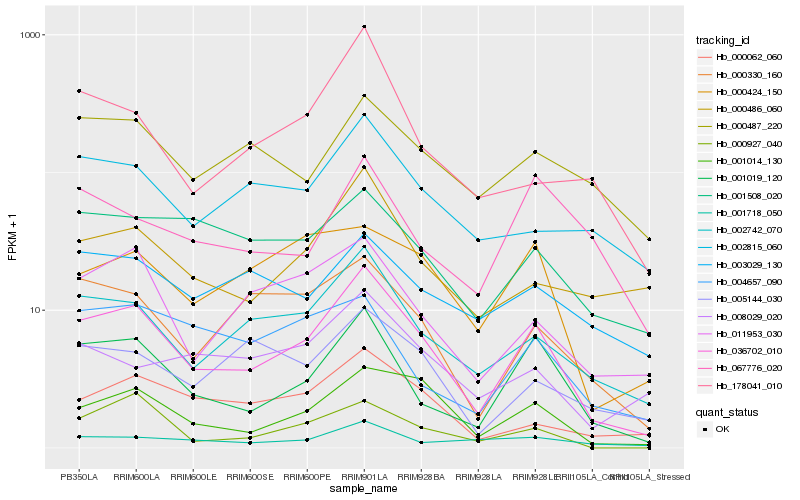
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_036702_010 |
0.0 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 2 |
Hb_001019_120 |
0.1365745831 |
- |
- |
PREDICTED: uncharacterized protein LOC105639315 [Jatropha curcas] |
| 3 |
Hb_001014_130 |
0.1474716669 |
- |
- |
PREDICTED: uncharacterized protein LOC105647301 [Jatropha curcas] |
| 4 |
Hb_002742_070 |
0.1483918957 |
- |
- |
PREDICTED: villin-4-like [Jatropha curcas] |
| 5 |
Hb_011953_030 |
0.1800331951 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0003s14450g [Populus trichocarpa] |
| 6 |
Hb_000330_160 |
0.1822492271 |
- |
- |
hypothetical protein JCGZ_04462 [Jatropha curcas] |
| 7 |
Hb_000062_060 |
0.1840266182 |
transcription factor |
TF Family: Orphans |
multi-sensor hybrid histidine kinase [Plautia stali symbiont] |
| 8 |
Hb_000927_040 |
0.1843004308 |
- |
- |
- |
| 9 |
Hb_000486_060 |
0.1915561962 |
- |
- |
PREDICTED: immediate early response 3-interacting protein 1-like isoform X2 [Glycine max] |
| 10 |
Hb_002815_060 |
0.1969887343 |
- |
- |
hypothetical protein B456_002G110500 [Gossypium raimondii] |
| 11 |
Hb_000424_150 |
0.1991396444 |
- |
- |
PREDICTED: copper transporter 5.1-like [Jatropha curcas] |
| 12 |
Hb_005144_030 |
0.2005802404 |
- |
- |
PREDICTED: alpha-L-fucosidase 1 [Jatropha curcas] |
| 13 |
Hb_001508_020 |
0.2009165666 |
- |
- |
myosin vIII, putative [Ricinus communis] |
| 14 |
Hb_178041_010 |
0.2019812996 |
- |
- |
PREDICTED: signal peptide peptidase-like 2 [Populus euphratica] |
| 15 |
Hb_000487_220 |
0.2042455403 |
- |
- |
eukaryotic translation elongation factor, putative [Ricinus communis] |
| 16 |
Hb_001718_050 |
0.2059011371 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Brassica rapa] |
| 17 |
Hb_008029_020 |
0.2100256588 |
- |
- |
PREDICTED: uncharacterized protein LOC105631777 [Jatropha curcas] |
| 18 |
Hb_003029_130 |
0.2122408017 |
- |
- |
carboxypeptidase regulatory region-containingprotein, putative [Ricinus communis] |
| 19 |
Hb_004657_090 |
0.2128647952 |
- |
- |
Chromosome-associated kinesin KLP1, putative [Ricinus communis] |
| 20 |
Hb_067776_020 |
0.2133011493 |
- |
- |
PREDICTED: signal recognition particle 54 kDa protein, chloroplastic [Jatropha curcas] |