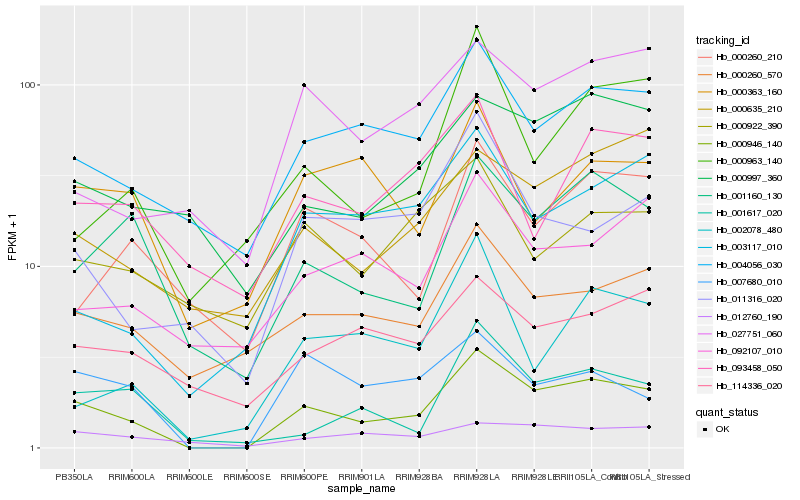
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000946_140 |
0.0 |
- |
- |
- |
| 2 |
Hb_004056_030 |
0.182120531 |
- |
- |
hypothetical protein 33 [Hevea brasiliensis] |
| 3 |
Hb_001160_130 |
0.1883765571 |
- |
- |
Glycolipid transfer protein, putative [Ricinus communis] |
| 4 |
Hb_007680_010 |
0.1928950557 |
- |
- |
hypothetical protein JCGZ_09539 [Jatropha curcas] |
| 5 |
Hb_000260_570 |
0.2017455768 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_001617_020 |
0.2062310622 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 7 |
Hb_000997_360 |
0.2065405399 |
- |
- |
hypothetical protein AALP_AAs70636U000100 [Arabis alpina] |
| 8 |
Hb_000635_210 |
0.2093485129 |
- |
- |
unknown [Populus trichocarpa] |
| 9 |
Hb_011316_020 |
0.2105124014 |
- |
- |
PREDICTED: uncharacterized protein LOC105648558 [Jatropha curcas] |
| 10 |
Hb_012760_190 |
0.2108533967 |
- |
- |
PREDICTED: fatty acyl-CoA reductase 2 [Jatropha curcas] |
| 11 |
Hb_114336_020 |
0.2137202491 |
- |
- |
PREDICTED: cytosolic Fe-S cluster assembly factor narfl [Fragaria vesca subsp. vesca] |
| 12 |
Hb_000363_160 |
0.2143696057 |
- |
- |
Heterogeneous nuclear ribonucleoprotein 27C, putative [Ricinus communis] |
| 13 |
Hb_092107_010 |
0.2160786478 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_027751_060 |
0.2164640339 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10-B [Jatropha curcas] |
| 15 |
Hb_093458_050 |
0.2182823419 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 16 |
Hb_002078_480 |
0.2205870907 |
- |
- |
hypothetical protein CICLE_v10033318mg [Citrus clementina] |
| 17 |
Hb_003117_010 |
0.220667847 |
- |
- |
PREDICTED: uncharacterized protein LOC100259103 isoform X1 [Vitis vinifera] |
| 18 |
Hb_000963_140 |
0.2214775347 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 19 |
Hb_000260_210 |
0.2215700967 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000922_390 |
0.2224889918 |
- |
- |
PREDICTED: uncharacterized protein LOC105640364 [Jatropha curcas] |