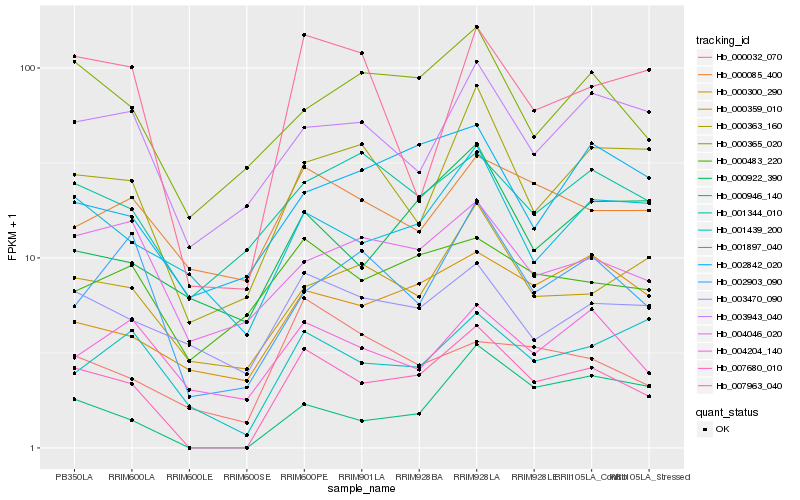
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007680_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_09539 [Jatropha curcas] |
| 2 |
Hb_000946_140 |
0.1928950557 |
- |
- |
- |
| 3 |
Hb_000363_160 |
0.2007713935 |
- |
- |
Heterogeneous nuclear ribonucleoprotein 27C, putative [Ricinus communis] |
| 4 |
Hb_007963_040 |
0.2025178318 |
- |
- |
dopamine beta-monooxygenase, putative [Ricinus communis] |
| 5 |
Hb_001897_040 |
0.2119063248 |
- |
- |
26S proteasome non-atpase regulatory subunit, putative [Ricinus communis] |
| 6 |
Hb_000365_020 |
0.2129202497 |
- |
- |
Vacuolar sorting receptor 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_002903_090 |
0.2171484133 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 8 |
Hb_000359_010 |
0.2176832932 |
- |
- |
o-linked n-acetylglucosamine transferase, ogt, putative [Ricinus communis] |
| 9 |
Hb_000300_290 |
0.218213027 |
- |
- |
PREDICTED: uncharacterized protein LOC105632583 [Jatropha curcas] |
| 10 |
Hb_003943_040 |
0.2202615892 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_003470_090 |
0.2218371762 |
- |
- |
PREDICTED: pumilio homolog 12 [Jatropha curcas] |
| 12 |
Hb_001439_200 |
0.2220519602 |
- |
- |
Transmembrane protein Tmp21 precursor, putative [Ricinus communis] |
| 13 |
Hb_000032_070 |
0.2226518122 |
- |
- |
hypothetical protein POPTR_0003s13760g [Populus trichocarpa] |
| 14 |
Hb_004046_020 |
0.2244511527 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g60570-like [Jatropha curcas] |
| 15 |
Hb_000922_390 |
0.2257494156 |
- |
- |
PREDICTED: uncharacterized protein LOC105640364 [Jatropha curcas] |
| 16 |
Hb_000483_220 |
0.2257782238 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2-like [Jatropha curcas] |
| 17 |
Hb_002842_020 |
0.2268238447 |
- |
- |
Prolactin regulatory element-binding protein, putative [Ricinus communis] |
| 18 |
Hb_000085_400 |
0.2283297493 |
- |
- |
Heterogeneous nuclear ribonucleoprotein 27C, putative [Ricinus communis] |
| 19 |
Hb_004204_140 |
0.2284851953 |
transcription factor |
TF Family: C2H2 |
hypothetical protein RCOM_1470470 [Ricinus communis] |
| 20 |
Hb_001344_010 |
0.2288236238 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI11 [Jatropha curcas] |