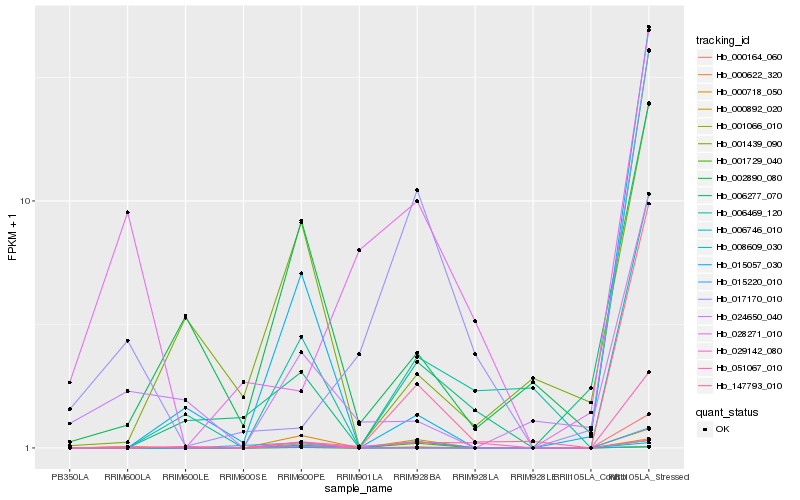
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000622_320 |
0.0 |
- |
- |
PREDICTED: chitinase 1-like [Vitis vinifera] |
| 2 |
Hb_029142_080 |
0.1877467012 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 3 |
Hb_000164_060 |
0.2526898839 |
- |
- |
- |
| 4 |
Hb_001066_010 |
0.273358082 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 5 |
Hb_024650_040 |
0.2742831474 |
- |
- |
PREDICTED: josephin-like protein [Jatropha curcas] |
| 6 |
Hb_002890_080 |
0.2898030585 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000892_020 |
0.2981391274 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_015057_030 |
0.3073953558 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-52, partial [Jatropha curcas] |
| 9 |
Hb_008609_030 |
0.3130249629 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 10 |
Hb_015220_010 |
0.3231113706 |
- |
- |
PREDICTED: ribonuclease 3-like protein 3 [Jatropha curcas] |
| 11 |
Hb_051067_010 |
0.3244856117 |
- |
- |
- |
| 12 |
Hb_000718_050 |
0.3268436946 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X2 [Populus euphratica] |
| 13 |
Hb_006746_010 |
0.3327551944 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 14 |
Hb_006469_120 |
0.3350953674 |
- |
- |
hypothetical protein POPTR_0005s16430g [Populus trichocarpa] |
| 15 |
Hb_006277_070 |
0.3368028013 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001439_090 |
0.3419751591 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_028271_010 |
0.3494256242 |
- |
- |
PREDICTED: trans-resveratrol di-O-methyltransferase-like [Populus euphratica] |
| 18 |
Hb_001729_040 |
0.3498370549 |
- |
- |
hypothetical protein glysoja_049826, partial [Glycine soja] |
| 19 |
Hb_147793_010 |
0.3518856506 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase PUB24-like [Jatropha curcas] |
| 20 |
Hb_017170_010 |
0.3556119121 |
- |
- |
PREDICTED: trans-resveratrol di-O-methyltransferase-like [Populus euphratica] |