| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
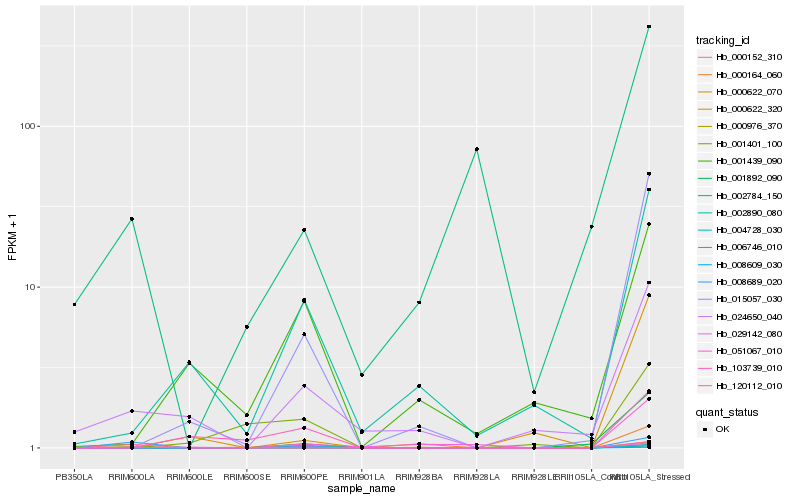
Hb_029142_080 |
0.0 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 2 |
Hb_000622_320 |
0.1877467012 |
- |
- |
PREDICTED: chitinase 1-like [Vitis vinifera] |
| 3 |
Hb_008609_030 |
0.2550198104 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 4 |
Hb_015057_030 |
0.27745154 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-52, partial [Jatropha curcas] |
| 5 |
Hb_024650_040 |
0.2800861794 |
- |
- |
PREDICTED: josephin-like protein [Jatropha curcas] |
| 6 |
Hb_006746_010 |
0.2803112671 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 7 |
Hb_002890_080 |
0.3060576153 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001892_090 |
0.3182714501 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000164_060 |
0.3536388481 |
- |
- |
- |
| 10 |
Hb_051067_010 |
0.3575074736 |
- |
- |
- |
| 11 |
Hb_004728_030 |
0.3580397101 |
- |
- |
hypothetical protein, partial [Lactobacillus brevis] |
| 12 |
Hb_001439_090 |
0.3582368599 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_008689_020 |
0.3603368582 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_120112_010 |
0.3622503218 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 15 |
Hb_001401_100 |
0.3628459812 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 16 |
Hb_103739_010 |
0.3639325964 |
- |
- |
sodium/hydrogen exchanger, putative [Ricinus communis] |
| 17 |
Hb_000152_310 |
0.3645561865 |
- |
- |
- |
| 18 |
Hb_000976_370 |
0.3646207248 |
- |
- |
beta-glucosidase, putative [Ricinus communis] |
| 19 |
Hb_000622_070 |
0.3669555901 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002784_150 |
0.3690382181 |
- |
- |
hypothetical protein POPTR_0001s06620g [Populus trichocarpa] |