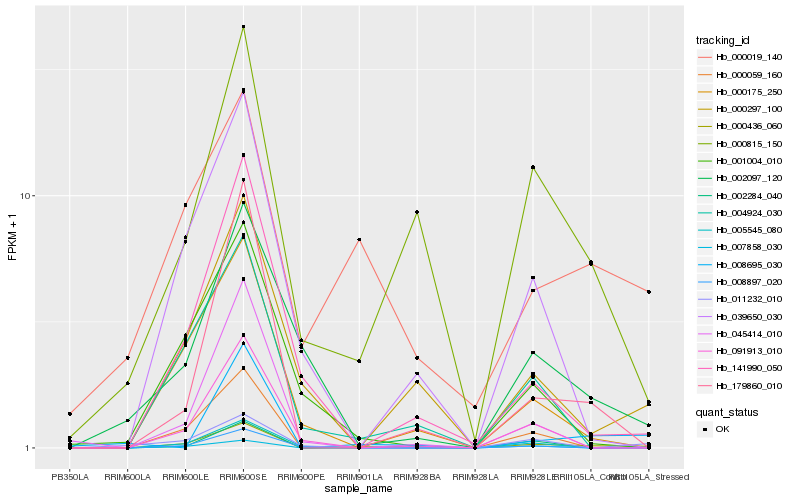
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000436_060 |
0.0 |
- |
- |
hypothetical protein VITISV_016971 [Vitis vinifera] |
| 2 |
Hb_000059_160 |
0.2366490525 |
- |
- |
hypothetical protein JCGZ_04974 [Jatropha curcas] |
| 3 |
Hb_179860_010 |
0.2577844825 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_007858_030 |
0.2639193974 |
- |
- |
retrotransposon protein, putative, Ty3-gypsy sub-class [Oryza sativa Japonica Group] |
| 5 |
Hb_005545_080 |
0.2662641506 |
- |
- |
hypothetical protein JCGZ_01293 [Jatropha curcas] |
| 6 |
Hb_000175_250 |
0.2766922624 |
- |
- |
PREDICTED: cucumber peeling cupredoxin-like [Jatropha curcas] |
| 7 |
Hb_141990_050 |
0.2800198021 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 8 |
Hb_004924_030 |
0.2830499196 |
- |
- |
- |
| 9 |
Hb_011232_010 |
0.2843771379 |
- |
- |
Leucine-rich repeat receptor-like protein kinase family protein [Theobroma cacao] |
| 10 |
Hb_091913_010 |
0.2874002568 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 11 |
Hb_008897_020 |
0.2885160393 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Musa acuminata subsp. malaccensis] |
| 12 |
Hb_000297_100 |
0.2888070321 |
transcription factor |
TF Family: NAC |
NAC transcription factor 038 [Jatropha curcas] |
| 13 |
Hb_001004_010 |
0.2897403804 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002284_040 |
0.2913936125 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_002097_120 |
0.2922744118 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 16 |
Hb_000815_150 |
0.295317641 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 17 |
Hb_000019_140 |
0.2979789315 |
- |
- |
Light-regulated protein precursor, putative [Ricinus communis] |
| 18 |
Hb_008695_030 |
0.2979800413 |
- |
- |
hypothetical protein JCGZ_12043 [Jatropha curcas] |
| 19 |
Hb_039650_030 |
0.2994826254 |
- |
- |
hypothetical protein RCOM_2128750 [Ricinus communis] |
| 20 |
Hb_045414_010 |
0.3017104297 |
- |
- |
unnamed protein product [Vitis vinifera] |