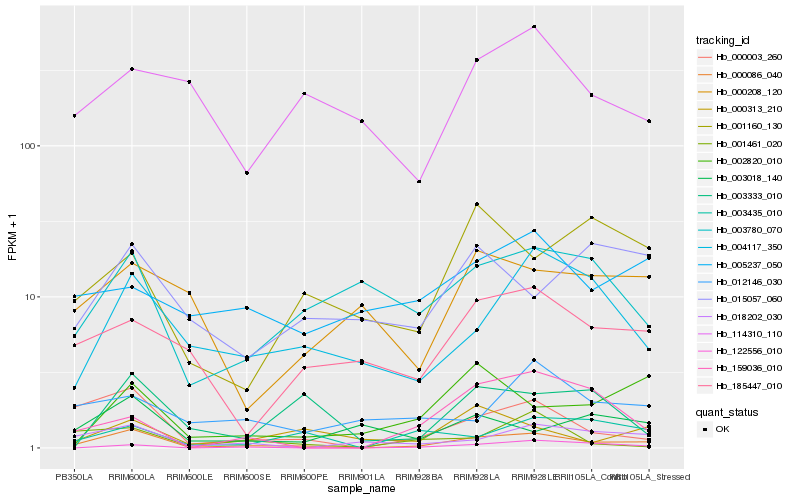
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000086_040 |
0.0 |
- |
- |
PREDICTED: legumin A-like [Jatropha curcas] |
| 2 |
Hb_000003_260 |
0.2115316859 |
- |
- |
- |
| 3 |
Hb_018202_030 |
0.2706068334 |
- |
- |
PREDICTED: uncharacterized protein LOC103419743 [Malus domestica] |
| 4 |
Hb_004117_350 |
0.2805428788 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP4 [Jatropha curcas] |
| 5 |
Hb_185447_010 |
0.2970159425 |
- |
- |
hypothetical protein JCGZ_13512 [Jatropha curcas] |
| 6 |
Hb_003435_010 |
0.299444226 |
- |
- |
PREDICTED: ras-related protein RABC2a-like [Jatropha curcas] |
| 7 |
Hb_003780_070 |
0.3003936011 |
- |
- |
PREDICTED: pirin-like protein [Jatropha curcas] |
| 8 |
Hb_122556_010 |
0.3047046765 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |
| 9 |
Hb_000313_210 |
0.3056117591 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002820_010 |
0.3094823251 |
- |
- |
PREDICTED: general transcription factor IIH subunit 3 isoform X2 [Jatropha curcas] |
| 11 |
Hb_003018_140 |
0.3107099958 |
- |
- |
PREDICTED: endoglucanase 24-like [Jatropha curcas] |
| 12 |
Hb_015057_060 |
0.316374361 |
- |
- |
- |
| 13 |
Hb_003333_010 |
0.3173921069 |
- |
- |
PREDICTED: uncharacterized protein LOC105645175 [Jatropha curcas] |
| 14 |
Hb_159036_010 |
0.3184134226 |
- |
- |
- |
| 15 |
Hb_012146_030 |
0.3185645848 |
- |
- |
- |
| 16 |
Hb_001461_020 |
0.3187597235 |
- |
- |
PREDICTED: uncharacterized protein LOC104901272 [Beta vulgaris subsp. vulgaris] |
| 17 |
Hb_114310_110 |
0.3200144112 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 18 |
Hb_001160_130 |
0.3207552124 |
- |
- |
Glycolipid transfer protein, putative [Ricinus communis] |
| 19 |
Hb_000208_120 |
0.32122991 |
- |
- |
PREDICTED: CAP-Gly domain-containing linker protein 1-like [Jatropha curcas] |
| 20 |
Hb_005237_050 |
0.3218280515 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105630924 [Jatropha curcas] |