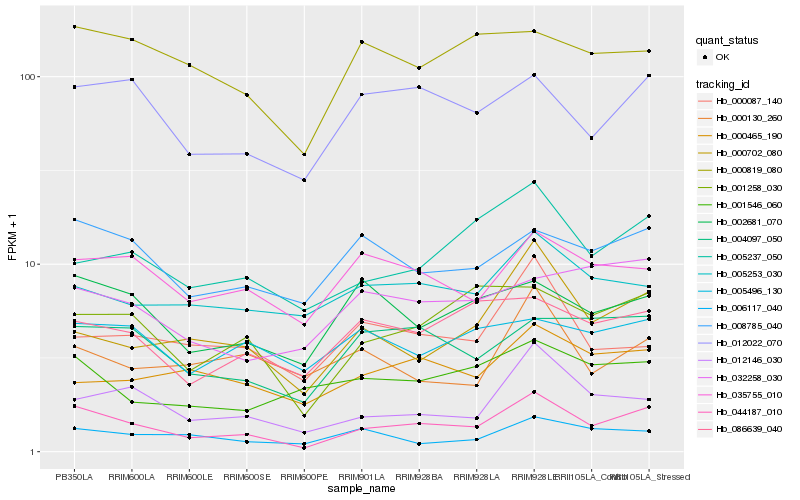
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_044187_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 2 |
Hb_001258_030 |
0.1509000384 |
- |
- |
tRNA-dihydrouridine synthase, putative [Ricinus communis] |
| 3 |
Hb_000702_080 |
0.1574859262 |
- |
- |
PREDICTED: uncharacterized protein LOC105630492 [Jatropha curcas] |
| 4 |
Hb_012022_070 |
0.1584488121 |
- |
- |
RecName: Full=Probable pyridoxal 5'-phosphate synthase subunit PDX1; Short=PLP synthase subunit PDX1; AltName: Full=Ethylene-inducible protein HEVER [Hevea brasiliensis] |
| 5 |
Hb_012146_030 |
0.1588769705 |
- |
- |
- |
| 6 |
Hb_004097_050 |
0.1595588241 |
- |
- |
PREDICTED: squalene monooxygenase-like [Jatropha curcas] |
| 7 |
Hb_005237_050 |
0.1597098392 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105630924 [Jatropha curcas] |
| 8 |
Hb_008785_040 |
0.1662510214 |
- |
- |
PREDICTED: DNA repair protein UVH3 [Jatropha curcas] |
| 9 |
Hb_002681_070 |
0.1704465015 |
- |
- |
PREDICTED: uncharacterized protein LOC105633833 isoform X1 [Jatropha curcas] |
| 10 |
Hb_086639_040 |
0.1719682709 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase gamma 7-like [Jatropha curcas] |
| 11 |
Hb_035755_010 |
0.1757451389 |
- |
- |
unknown [Populus trichocarpa] |
| 12 |
Hb_001546_060 |
0.1789816189 |
- |
- |
PREDICTED: uncharacterized protein LOC105133943 isoform X4 [Populus euphratica] |
| 13 |
Hb_000819_080 |
0.1823851451 |
- |
- |
Plastid-lipid-associated protein, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_006117_040 |
0.1831611996 |
- |
- |
PREDICTED: seed biotin-containing protein SBP65 [Jatropha curcas] |
| 15 |
Hb_032258_030 |
0.1832845479 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647563 [Jatropha curcas] |
| 16 |
Hb_000465_190 |
0.1834105767 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HOS1 isoform X2 [Jatropha curcas] |
| 17 |
Hb_005253_030 |
0.1845083364 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_000087_140 |
0.1850394266 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g27270 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000130_260 |
0.1893851882 |
rubber biosynthesis |
Gene Name: Dimethyallyltransferase |
PREDICTED: tRNA dimethylallyltransferase 9 isoform X1 [Jatropha curcas] |
| 20 |
Hb_005496_130 |
0.1905068491 |
- |
- |
PREDICTED: activating signal cointegrator 1 complex subunit 2 isoform X2 [Jatropha curcas] |