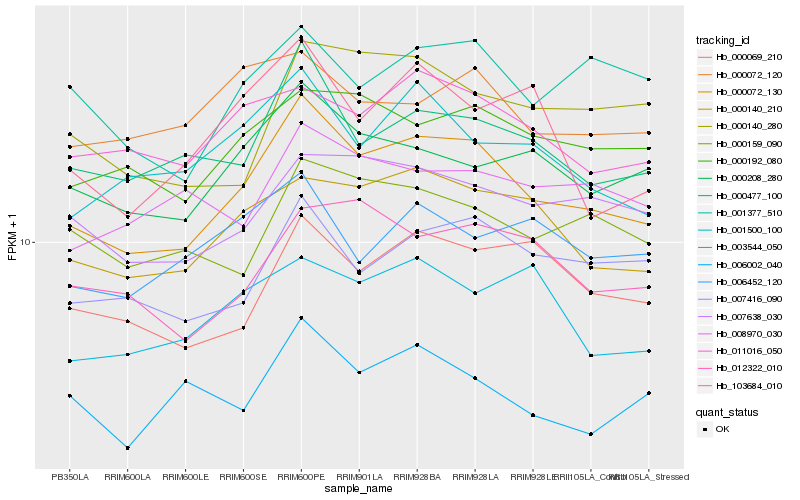
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000072_130 |
0.0 |
- |
- |
hypothetical protein OsJ_28567 [Oryza sativa Japonica Group] |
| 2 |
Hb_000140_210 |
0.0802302041 |
- |
- |
sec10, putative [Ricinus communis] |
| 3 |
Hb_000477_100 |
0.081572344 |
- |
- |
PREDICTED: GDP-mannose transporter GONST4 [Jatropha curcas] |
| 4 |
Hb_000208_280 |
0.0866921185 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 78 [Jatropha curcas] |
| 5 |
Hb_007638_030 |
0.0894925014 |
- |
- |
PREDICTED: phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN [Jatropha curcas] |
| 6 |
Hb_000192_080 |
0.0899251802 |
- |
- |
PREDICTED: serine/threonine-protein kinase SAPK3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_007416_090 |
0.0908044471 |
- |
- |
UDP-sugar transporter, putative [Ricinus communis] |
| 8 |
Hb_001500_100 |
0.0928955387 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000069_210 |
0.0930418395 |
- |
- |
PREDICTED: exportin-2 [Jatropha curcas] |
| 10 |
Hb_011016_050 |
0.0930840782 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI7 isoform X1 [Jatropha curcas] |
| 11 |
Hb_012322_010 |
0.0952694884 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 12 |
Hb_000159_090 |
0.0963963921 |
- |
- |
hypothetical protein POPTR_0015s153201g, partial [Populus trichocarpa] |
| 13 |
Hb_001377_510 |
0.1012802321 |
- |
- |
ormdl, putative [Ricinus communis] |
| 14 |
Hb_003544_050 |
0.1023684257 |
- |
- |
PREDICTED: uncharacterized protein LOC105644300 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000140_280 |
0.1029591573 |
- |
- |
hypothetical protein B456_001G157900 [Gossypium raimondii] |
| 16 |
Hb_008970_030 |
0.1037896808 |
- |
- |
arf gtpase-activating protein, putative [Ricinus communis] |
| 17 |
Hb_000072_120 |
0.10441863 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 18 |
Hb_103684_010 |
0.1045133806 |
- |
- |
PREDICTED: cullin-1 isoform X3 [Nicotiana tomentosiformis] |
| 19 |
Hb_006452_120 |
0.1051338026 |
- |
- |
PREDICTED: protein FAM188A [Jatropha curcas] |
| 20 |
Hb_006002_040 |
0.1053600238 |
- |
- |
PREDICTED: uncharacterized protein LOC105638085 [Jatropha curcas] |