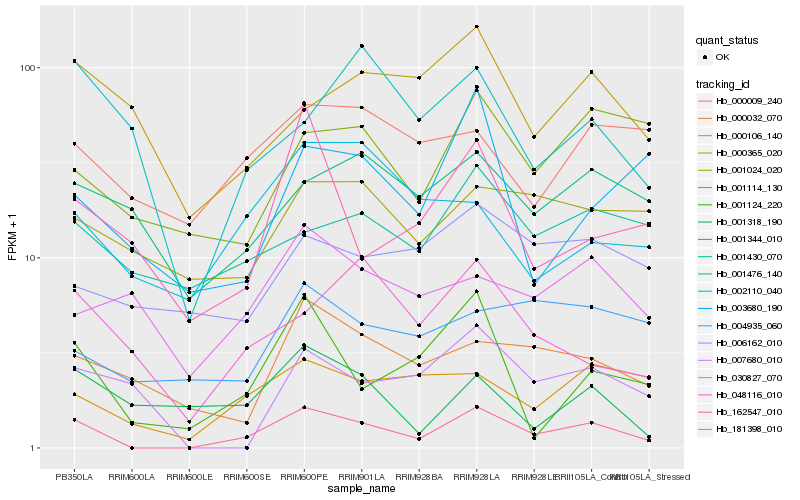
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_181398_010 |
0.0 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 2 |
Hb_001124_220 |
0.2526180589 |
- |
- |
DsRNA-binding protein 5, putative isoform 1 [Theobroma cacao] |
| 3 |
Hb_000106_140 |
0.271534328 |
- |
- |
- |
| 4 |
Hb_000032_070 |
0.2774536806 |
- |
- |
hypothetical protein POPTR_0003s13760g [Populus trichocarpa] |
| 5 |
Hb_048116_010 |
0.2799168999 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 6 |
Hb_162547_010 |
0.283758133 |
- |
- |
- |
| 7 |
Hb_001114_130 |
0.2853681939 |
transcription factor |
TF Family: MIKC |
PREDICTED: MADS-box protein SVP-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_001318_190 |
0.289242411 |
- |
- |
PREDICTED: uncharacterized protein LOC102617255 [Citrus sinensis] |
| 9 |
Hb_007680_010 |
0.2898695811 |
- |
- |
hypothetical protein JCGZ_09539 [Jatropha curcas] |
| 10 |
Hb_001430_070 |
0.2902941413 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001344_010 |
0.2910081099 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI11 [Jatropha curcas] |
| 12 |
Hb_003680_190 |
0.2919869296 |
- |
- |
hypothetical protein POPTR_0006s11880g [Populus trichocarpa] |
| 13 |
Hb_001476_140 |
0.2935789056 |
- |
- |
PREDICTED: uncharacterized protein LOC105630497 [Jatropha curcas] |
| 14 |
Hb_030827_070 |
0.2950897572 |
- |
- |
PREDICTED: transmembrane protein 161B [Jatropha curcas] |
| 15 |
Hb_001024_020 |
0.2973459809 |
- |
- |
hypothetical protein JCGZ_08989 [Jatropha curcas] |
| 16 |
Hb_002110_040 |
0.2980273088 |
- |
- |
hypothetical protein AMTR_s00135p00074190 [Amborella trichopoda] |
| 17 |
Hb_004935_060 |
0.298308831 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 18 |
Hb_000365_020 |
0.2988495213 |
- |
- |
Vacuolar sorting receptor 1 precursor, putative [Ricinus communis] |
| 19 |
Hb_000009_240 |
0.2999377871 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF185-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_006162_010 |
0.3014148571 |
- |
- |
CAZy families GH95 protein, partial [uncultured Chitinophaga sp.] |