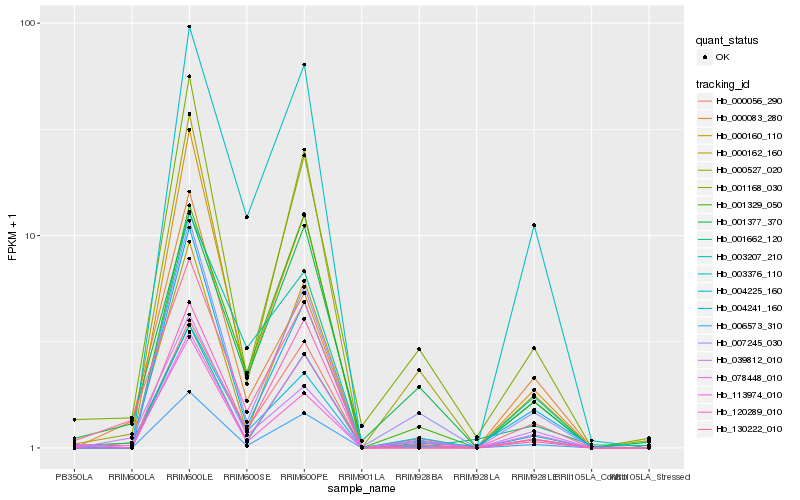
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_130222_010 |
0.0 |
- |
- |
hypothetical protein RCOM_1272620 [Ricinus communis] |
| 2 |
Hb_113974_010 |
0.1321380388 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_000056_290 |
0.1467826121 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000162_160 |
0.1475425996 |
- |
- |
PREDICTED: auxin transporter-like protein 5 [Populus euphratica] |
| 5 |
Hb_078448_010 |
0.1508256528 |
- |
- |
hypothetical protein L484_017526 [Morus notabilis] |
| 6 |
Hb_006573_310 |
0.1535880585 |
- |
- |
Tellurite resistance protein tehA, putative [Ricinus communis] |
| 7 |
Hb_000083_280 |
0.1559437282 |
- |
- |
copper ion binding protein, putative [Ricinus communis] |
| 8 |
Hb_039812_010 |
0.156478367 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 9 |
Hb_001377_370 |
0.1580383299 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 10 |
Hb_000527_020 |
0.1594658234 |
- |
- |
PREDICTED: uncharacterized protein LOC105649490 [Jatropha curcas] |
| 11 |
Hb_001168_030 |
0.1601957421 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin S13-7 [Jatropha curcas] |
| 12 |
Hb_003376_110 |
0.1607771314 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_007245_030 |
0.1641768362 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000160_110 |
0.1664743331 |
- |
- |
PREDICTED: uncharacterized protein LOC100253584 [Vitis vinifera] |
| 15 |
Hb_004225_160 |
0.1674587548 |
- |
- |
PREDICTED: kiwellin-like [Jatropha curcas] |
| 16 |
Hb_001329_050 |
0.1681478943 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_120289_010 |
0.1683545496 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Jatropha curcas] |
| 18 |
Hb_003207_210 |
0.1696394908 |
- |
- |
PREDICTED: uncharacterized protein LOC105645885 [Jatropha curcas] |
| 19 |
Hb_004241_160 |
0.1698200463 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase SD1-7 [Jatropha curcas] |
| 20 |
Hb_001662_120 |
0.170256794 |
- |
- |
PREDICTED: ammonium transporter 3 member 1-like [Jatropha curcas] |