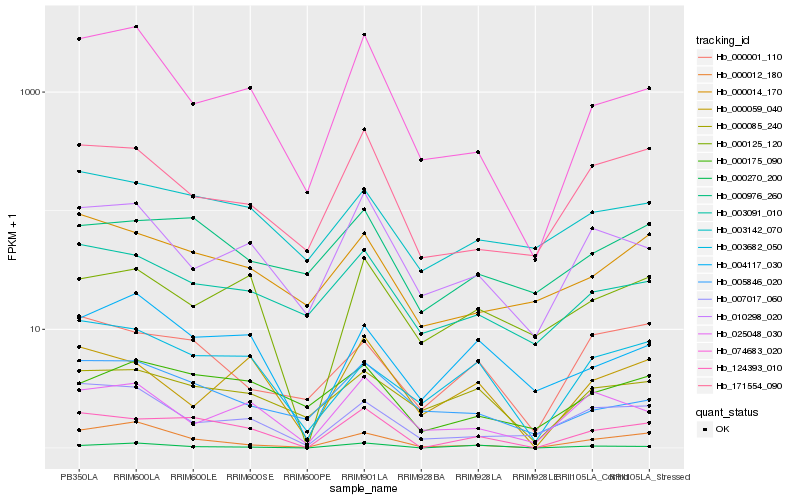
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_124393_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_000059_040 |
0.2240752723 |
- |
- |
Uncharacterized protein TCM_020504 [Theobroma cacao] |
| 3 |
Hb_074683_020 |
0.2264004022 |
- |
- |
PREDICTED: 60S ribosomal protein L27-3 [Jatropha curcas] |
| 4 |
Hb_003682_050 |
0.228327704 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 5 |
Hb_171554_090 |
0.2322134141 |
- |
- |
PREDICTED: 10 kDa chaperonin [Jatropha curcas] |
| 6 |
Hb_000012_180 |
0.2334905363 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000125_120 |
0.2351286925 |
transcription factor |
TF Family: mTERF |
- |
| 8 |
Hb_000270_200 |
0.2352910483 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_000175_090 |
0.2370502326 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000001_110 |
0.2376861637 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_010298_020 |
0.2385626447 |
- |
- |
PREDICTED: uncharacterized protein At2g02148 isoform X4 [Vitis vinifera] |
| 12 |
Hb_007017_060 |
0.2402281096 |
- |
- |
PREDICTED: 5-formyltetrahydrofolate cyclo-ligase, mitochondrial-like [Jatropha curcas] |
| 13 |
Hb_000976_260 |
0.2408074333 |
- |
- |
- |
| 14 |
Hb_000085_240 |
0.2409320882 |
- |
- |
fimbrin, putative [Ricinus communis] |
| 15 |
Hb_000014_170 |
0.24289054 |
- |
- |
hypothetical protein CISIN_1g0379742mg, partial [Citrus sinensis] |
| 16 |
Hb_003091_010 |
0.2430823053 |
- |
- |
PREDICTED: uncharacterized protein LOC105131887 [Populus euphratica] |
| 17 |
Hb_004117_030 |
0.243179485 |
- |
- |
PREDICTED: uncharacterized protein LOC104449809 [Eucalyptus grandis] |
| 18 |
Hb_003142_070 |
0.2436340964 |
- |
- |
hypothetical protein F383_14657 [Gossypium arboreum] |
| 19 |
Hb_025048_030 |
0.2458607857 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 20 |
Hb_005846_020 |
0.2463182522 |
- |
- |
PREDICTED: tubulin-folding cofactor C [Jatropha curcas] |