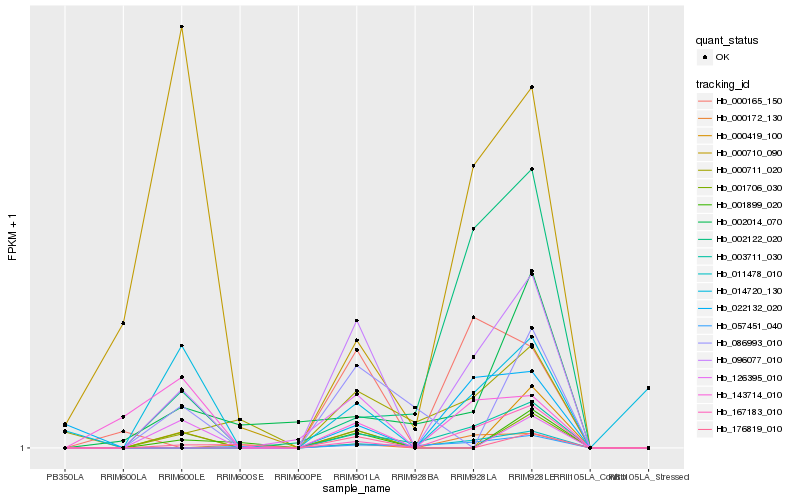
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_096077_010 |
0.0 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |
| 2 |
Hb_003711_030 |
0.3249870437 |
- |
- |
PREDICTED: uncharacterized protein LOC102621950 [Citrus sinensis] |
| 3 |
Hb_001706_030 |
0.3349515598 |
- |
- |
PREDICTED: uncharacterized protein LOC104880364 [Vitis vinifera] |
| 4 |
Hb_000165_150 |
0.3355895818 |
- |
- |
PREDICTED: pistil-specific extensin-like protein [Jatropha curcas] |
| 5 |
Hb_011478_010 |
0.3386919162 |
- |
- |
- |
| 6 |
Hb_167183_010 |
0.3431834007 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 7 |
Hb_002122_020 |
0.3538298835 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Jatropha curcas] |
| 8 |
Hb_000419_100 |
0.3570908666 |
- |
- |
- |
| 9 |
Hb_000711_020 |
0.36212868 |
- |
- |
major allergen Pru ar 1-like [Jatropha curcas] |
| 10 |
Hb_001899_020 |
0.3837293524 |
- |
- |
PREDICTED: phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase TPTE2 [Jatropha curcas] |
| 11 |
Hb_176819_010 |
0.3870250413 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 12 |
Hb_143714_010 |
0.3876641513 |
- |
- |
PREDICTED: phenolic glucoside malonyltransferase 1-like [Populus euphratica] |
| 13 |
Hb_000172_130 |
0.393683089 |
- |
- |
hypothetical protein JCGZ_24076 [Jatropha curcas] |
| 14 |
Hb_014720_130 |
0.3944508276 |
- |
- |
PREDICTED: uncharacterized protein LOC105640368 [Jatropha curcas] |
| 15 |
Hb_002014_070 |
0.4008255391 |
- |
- |
PREDICTED: probable chlorophyll(ide) b reductase NYC1, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_086993_010 |
0.4018938427 |
- |
- |
- |
| 17 |
Hb_057451_040 |
0.4028666464 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 18 |
Hb_022132_020 |
0.4050399147 |
- |
- |
hypothetical protein TRIUR3_12476 [Triticum urartu] |
| 19 |
Hb_000710_090 |
0.4074679208 |
- |
- |
PREDICTED: uncharacterized protein LOC104583249 [Brachypodium distachyon] |
| 20 |
Hb_126395_010 |
0.4086473264 |
- |
- |
hypothetical protein VITISV_043980 [Vitis vinifera] |