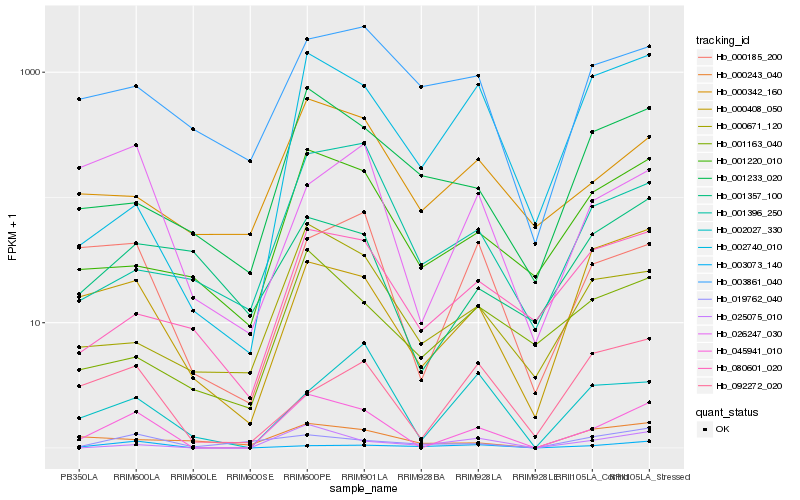
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_045941_010 |
0.0 |
- |
- |
PREDICTED: abscisic stress-ripening protein 2-like [Eucalyptus grandis] |
| 2 |
Hb_000185_200 |
0.2679479552 |
- |
- |
PREDICTED: protein trichome birefringence-like 42 [Jatropha curcas] |
| 3 |
Hb_000408_050 |
0.2708778366 |
- |
- |
hypothetical protein POPTR_0018s11350g [Populus trichocarpa] |
| 4 |
Hb_025075_010 |
0.2770910509 |
- |
- |
50S ribosomal protein L24 [Glycine soja] |
| 5 |
Hb_001220_010 |
0.2905532764 |
- |
- |
hypothetical protein JCGZ_00411 [Jatropha curcas] |
| 6 |
Hb_000671_120 |
0.2927394351 |
- |
- |
hypothetical protein JCGZ_23777 [Jatropha curcas] |
| 7 |
Hb_000342_160 |
0.2959380513 |
- |
- |
PREDICTED: sarcoplasmic reticulum histidine-rich calcium-binding protein [Jatropha curcas] |
| 8 |
Hb_003073_140 |
0.2975797328 |
- |
- |
F-box family protein [Theobroma cacao] |
| 9 |
Hb_026247_030 |
0.2982023994 |
- |
- |
PREDICTED: protein trichome birefringence-like 42 [Jatropha curcas] |
| 10 |
Hb_001233_020 |
0.299534012 |
- |
- |
hypothetical protein JCGZ_04109 [Jatropha curcas] |
| 11 |
Hb_080601_020 |
0.3031214035 |
- |
- |
PREDICTED: cytochrome c oxidase copper chaperone 1 [Jatropha curcas] |
| 12 |
Hb_002027_330 |
0.3083985256 |
- |
- |
- |
| 13 |
Hb_001163_040 |
0.3092792282 |
- |
- |
Cytochrome-c oxidases,electron carriers [Theobroma cacao] |
| 14 |
Hb_001396_250 |
0.3135523665 |
- |
- |
PREDICTED: stress-associated endoplasmic reticulum protein 2 [Jatropha curcas] |
| 15 |
Hb_019762_040 |
0.3137024093 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003861_040 |
0.3139248883 |
- |
- |
hypothetical protein JCGZ_16325 [Jatropha curcas] |
| 17 |
Hb_002740_010 |
0.3148468657 |
- |
- |
stress-induced hydrophobic peptide [Cleistogenes songorica] |
| 18 |
Hb_000243_040 |
0.3149659525 |
- |
- |
- |
| 19 |
Hb_001357_100 |
0.3151322509 |
- |
- |
PREDICTED: small ubiquitin-related modifier 1 [Jatropha curcas] |
| 20 |
Hb_092272_020 |
0.3157762066 |
- |
- |
hypothetical protein EUGRSUZ_F00278 [Eucalyptus grandis] |