| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
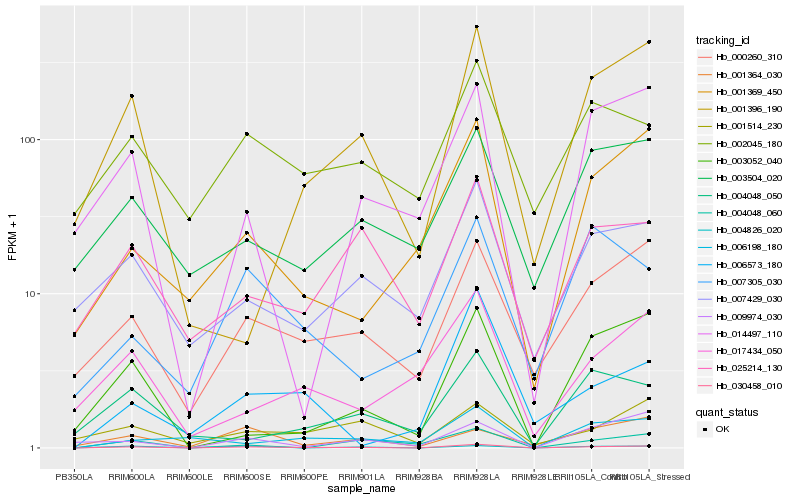
Hb_030458_010 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 2 |
Hb_004826_020 |
0.2254880248 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 3 |
Hb_000260_310 |
0.2346307675 |
- |
- |
PREDICTED: uncharacterized protein LOC105649060 [Jatropha curcas] |
| 4 |
Hb_003052_040 |
0.2607420977 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_007429_030 |
0.2653901207 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004048_060 |
0.2681667549 |
- |
- |
hypothetical protein PRUPE_ppa023788mg, partial [Prunus persica] |
| 7 |
Hb_025214_130 |
0.2703550081 |
- |
- |
PREDICTED: uncharacterized protein LOC105640854 [Jatropha curcas] |
| 8 |
Hb_001396_190 |
0.2709476572 |
- |
- |
PREDICTED: uncharacterized protein LOC105123025 [Populus euphratica] |
| 9 |
Hb_001369_450 |
0.2720726002 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 10 |
Hb_002045_180 |
0.2746468423 |
- |
- |
PREDICTED: uncharacterized protein LOC105649915 [Jatropha curcas] |
| 11 |
Hb_001514_230 |
0.2756055993 |
- |
- |
PREDICTED: pumilio homolog 12-like [Jatropha curcas] |
| 12 |
Hb_009974_030 |
0.2761816338 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g18390, mitochondrial-like [Jatropha curcas] |
| 13 |
Hb_001364_030 |
0.2765306752 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 14 |
Hb_014497_110 |
0.2779991679 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 25-like [Jatropha curcas] |
| 15 |
Hb_006573_180 |
0.2803913667 |
transcription factor |
TF Family: WRKY |
hypothetical protein RCOM_1581940 [Ricinus communis] |
| 16 |
Hb_007305_030 |
0.2835012217 |
- |
- |
serine-threonine protein kinase, putative [Ricinus communis] |
| 17 |
Hb_004048_050 |
0.2835929773 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570-like [Citrus sinensis] |
| 18 |
Hb_003504_020 |
0.2862880506 |
- |
- |
PREDICTED: uncharacterized protein LOC105647732 [Jatropha curcas] |
| 19 |
Hb_017434_050 |
0.2863596964 |
- |
- |
PREDICTED: importin subunit beta-3-like [Jatropha curcas] |
| 20 |
Hb_006198_180 |
0.2864168213 |
- |
- |
PREDICTED: thiamine-repressible mitochondrial transport protein THI74 isoform X1 [Jatropha curcas] |