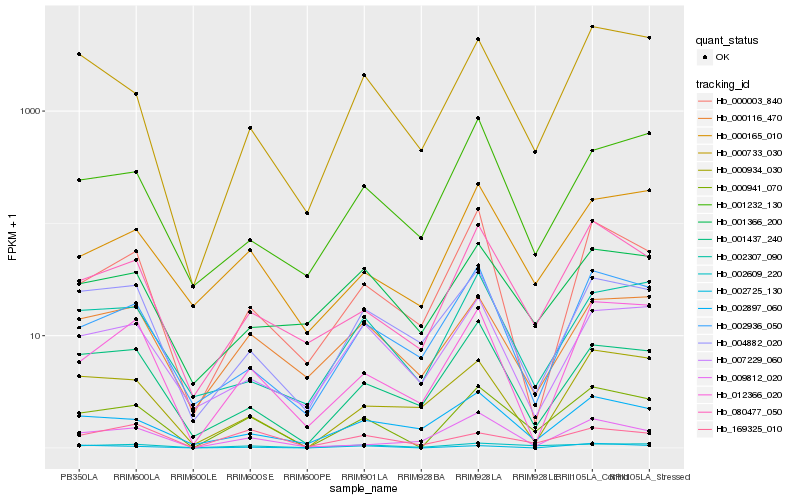
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000941_070 |
0.0 |
- |
- |
- |
| 2 |
Hb_080477_050 |
0.1819023002 |
- |
- |
hypothetical protein M569_16374 [Genlisea aurea] |
| 3 |
Hb_002609_220 |
0.1911184691 |
- |
- |
Leucine-rich repeat protein kinase family protein, putative [Theobroma cacao] |
| 4 |
Hb_000733_030 |
0.1944232966 |
- |
- |
PREDICTED: 18.1 kDa class I heat shock protein-like [Populus euphratica] |
| 5 |
Hb_000165_010 |
0.2007466721 |
- |
- |
hypothetical protein POPTR_0010s19120g [Populus trichocarpa] |
| 6 |
Hb_001437_240 |
0.2019123252 |
- |
- |
- |
| 7 |
Hb_012366_020 |
0.2031852909 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_000116_470 |
0.2032735974 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 10-like [Jatropha curcas] |
| 9 |
Hb_002936_050 |
0.20687754 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR18 isoform X2 [Jatropha curcas] |
| 10 |
Hb_004882_020 |
0.2073597429 |
- |
- |
- |
| 11 |
Hb_002307_090 |
0.2097470308 |
- |
- |
magnesium transporter CorA-like family protein [Populus trichocarpa] |
| 12 |
Hb_000003_840 |
0.2138272471 |
- |
- |
multidrug/pheromone exporter protein [Hevea brasiliensis] |
| 13 |
Hb_002725_130 |
0.2139171964 |
- |
- |
Osmotic avoidance abnormal protein, putative [Ricinus communis] |
| 14 |
Hb_002897_060 |
0.2144072819 |
- |
- |
PREDICTED: uncharacterized protein LOC105649233 [Jatropha curcas] |
| 15 |
Hb_169325_010 |
0.2149951781 |
- |
- |
PREDICTED: uncharacterized protein LOC103699787, partial [Phoenix dactylifera] |
| 16 |
Hb_009812_020 |
0.2193025477 |
- |
- |
- |
| 17 |
Hb_007229_060 |
0.22010677 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g44880 [Jatropha curcas] |
| 18 |
Hb_001232_130 |
0.2219005007 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL1-like [Jatropha curcas] |
| 19 |
Hb_000934_030 |
0.2221625171 |
- |
- |
- |
| 20 |
Hb_001366_200 |
0.2229024428 |
- |
- |
conserved hypothetical protein [Ricinus communis] |