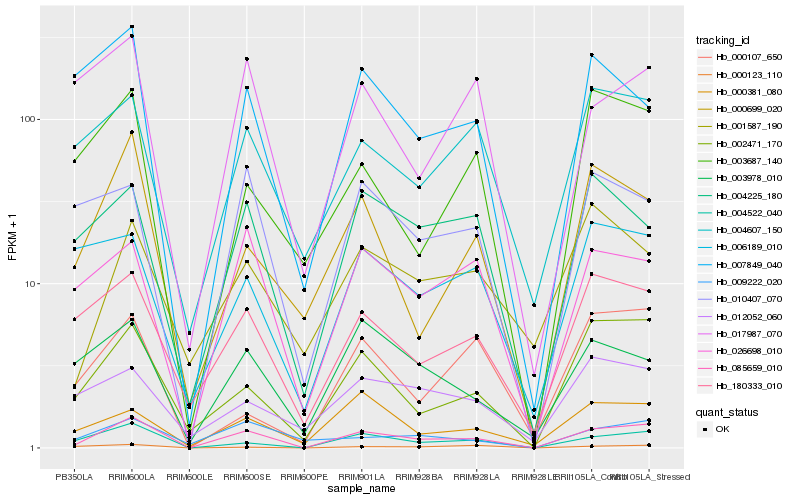
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_085659_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104216799 [Nicotiana sylvestris] |
| 2 |
Hb_004522_040 |
0.1684692835 |
- |
- |
PREDICTED: probable galacturonosyltransferase 9 [Populus euphratica] |
| 3 |
Hb_002471_170 |
0.1896205113 |
- |
- |
- |
| 4 |
Hb_026698_010 |
0.2054598431 |
- |
- |
2,4-dienoyl-CoA reductase, putative [Ricinus communis] |
| 5 |
Hb_180333_010 |
0.2059367244 |
- |
- |
- |
| 6 |
Hb_004225_180 |
0.2152107618 |
- |
- |
bel1 homeotic protein, putative [Ricinus communis] |
| 7 |
Hb_000381_080 |
0.2182347463 |
transcription factor |
TF Family: SNF2 |
PREDICTED: DNA repair protein RAD16 [Jatropha curcas] |
| 8 |
Hb_000699_020 |
0.2187730308 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase kinase 2-like [Jatropha curcas] |
| 9 |
Hb_004607_150 |
0.2194367078 |
- |
- |
PREDICTED: serine/threonine-protein kinase UCNL [Jatropha curcas] |
| 10 |
Hb_010407_070 |
0.2248355819 |
- |
- |
hypothetical protein POPTR_0008s16750g [Populus trichocarpa] |
| 11 |
Hb_003978_010 |
0.2274424171 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 12 |
Hb_007849_040 |
0.2303693928 |
- |
- |
PREDICTED: protein LIGHT-DEPENDENT SHORT HYPOCOTYLS 1-like [Vitis vinifera] |
| 13 |
Hb_009222_020 |
0.2317751763 |
- |
- |
hypothetical protein AALP_AA5G087000 [Arabis alpina] |
| 14 |
Hb_017987_070 |
0.2323489417 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT22-like [Jatropha curcas] |
| 15 |
Hb_001587_190 |
0.235947625 |
- |
- |
PREDICTED: uncharacterized protein LOC105762101 [Gossypium raimondii] |
| 16 |
Hb_003687_140 |
0.236551246 |
- |
- |
BnaCnng65050D [Brassica napus] |
| 17 |
Hb_000123_110 |
0.2369689021 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g39620-like [Pyrus x bretschneideri] |
| 18 |
Hb_006189_010 |
0.2412130134 |
- |
- |
- |
| 19 |
Hb_000107_650 |
0.2416518891 |
- |
- |
PREDICTED: cyclic phosphodiesterase-like [Jatropha curcas] |
| 20 |
Hb_012052_060 |
0.2418813597 |
- |
- |
hypothetical protein POPTR_0018s10190g [Populus trichocarpa] |