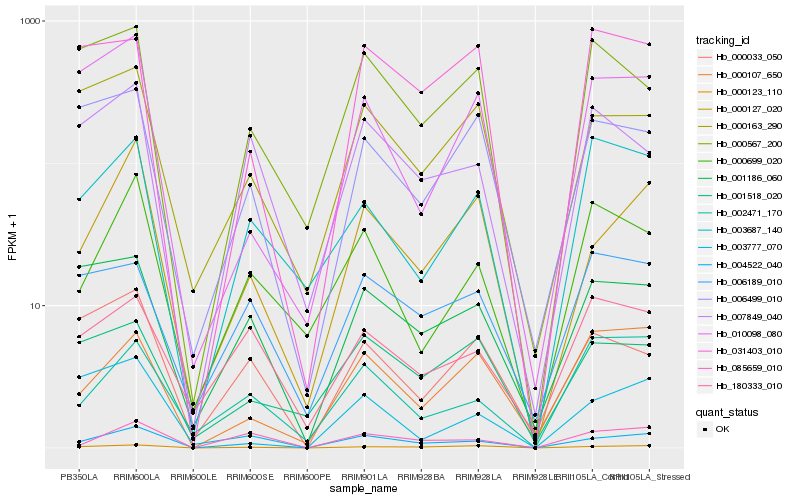
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004522_040 |
0.0 |
- |
- |
PREDICTED: probable galacturonosyltransferase 9 [Populus euphratica] |
| 2 |
Hb_000127_020 |
0.146441673 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 3 |
Hb_000123_110 |
0.1505184933 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g39620-like [Pyrus x bretschneideri] |
| 4 |
Hb_085659_010 |
0.1684692835 |
- |
- |
PREDICTED: uncharacterized protein LOC104216799 [Nicotiana sylvestris] |
| 5 |
Hb_001186_060 |
0.1795463821 |
- |
- |
- |
| 6 |
Hb_000107_650 |
0.1802961509 |
- |
- |
PREDICTED: cyclic phosphodiesterase-like [Jatropha curcas] |
| 7 |
Hb_000163_290 |
0.1845593241 |
- |
- |
PREDICTED: nucleolar protein 16 [Jatropha curcas] |
| 8 |
Hb_007849_040 |
0.1889499371 |
- |
- |
PREDICTED: protein LIGHT-DEPENDENT SHORT HYPOCOTYLS 1-like [Vitis vinifera] |
| 9 |
Hb_002471_170 |
0.1908685965 |
- |
- |
- |
| 10 |
Hb_001518_020 |
0.1943701208 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 13-like isoform X2 [Fragaria vesca subsp. vesca] |
| 11 |
Hb_006189_010 |
0.1968647915 |
- |
- |
- |
| 12 |
Hb_000699_020 |
0.1970899073 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase kinase 2-like [Jatropha curcas] |
| 13 |
Hb_031403_010 |
0.1972124182 |
- |
- |
hypothetical protein POPTR_0001s40140g [Populus trichocarpa] |
| 14 |
Hb_006499_010 |
0.2004095806 |
- |
- |
- |
| 15 |
Hb_010098_080 |
0.2005100432 |
- |
- |
PREDICTED: non-specific phospholipase C4-like [Jatropha curcas] |
| 16 |
Hb_000567_200 |
0.2011902763 |
- |
- |
PREDICTED: protein LIGHT-DEPENDENT SHORT HYPOCOTYLS 10 [Jatropha curcas] |
| 17 |
Hb_003777_070 |
0.2018570342 |
- |
- |
- |
| 18 |
Hb_180333_010 |
0.2033709219 |
- |
- |
- |
| 19 |
Hb_003687_140 |
0.2039857609 |
- |
- |
BnaCnng65050D [Brassica napus] |
| 20 |
Hb_000033_050 |
0.2065466107 |
- |
- |
- |