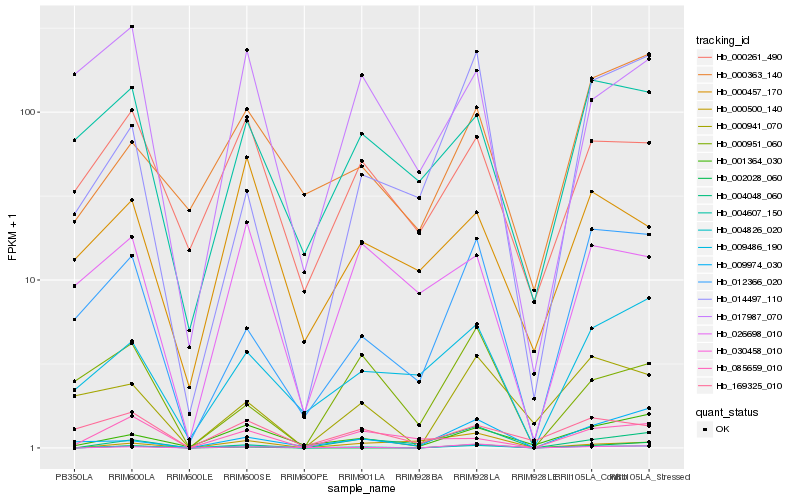
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004826_020 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 2 |
Hb_030458_010 |
0.2254880248 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 3 |
Hb_001364_030 |
0.2404743826 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 4 |
Hb_004048_060 |
0.283020828 |
- |
- |
hypothetical protein PRUPE_ppa023788mg, partial [Prunus persica] |
| 5 |
Hb_012366_020 |
0.293689058 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_085659_010 |
0.2941750849 |
- |
- |
PREDICTED: uncharacterized protein LOC104216799 [Nicotiana sylvestris] |
| 7 |
Hb_000941_070 |
0.2968621755 |
- |
- |
- |
| 8 |
Hb_000261_490 |
0.2975007998 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 5 [Jatropha curcas] |
| 9 |
Hb_009486_190 |
0.2984770573 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_014497_110 |
0.3022076407 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 25-like [Jatropha curcas] |
| 11 |
Hb_026698_010 |
0.3099037016 |
- |
- |
2,4-dienoyl-CoA reductase, putative [Ricinus communis] |
| 12 |
Hb_009974_030 |
0.3119042138 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g18390, mitochondrial-like [Jatropha curcas] |
| 13 |
Hb_169325_010 |
0.3124454102 |
- |
- |
PREDICTED: uncharacterized protein LOC103699787, partial [Phoenix dactylifera] |
| 14 |
Hb_002028_060 |
0.3145584848 |
- |
- |
- |
| 15 |
Hb_000500_140 |
0.3156644396 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 4 [Jatropha curcas] |
| 16 |
Hb_000457_170 |
0.316049936 |
- |
- |
PREDICTED: uncharacterized protein LOC105633066 [Jatropha curcas] |
| 17 |
Hb_004607_150 |
0.3182847622 |
- |
- |
PREDICTED: serine/threonine-protein kinase UCNL [Jatropha curcas] |
| 18 |
Hb_000363_140 |
0.3193685362 |
- |
- |
hypothetical protein CICLE_v10009052mg [Citrus clementina] |
| 19 |
Hb_017987_070 |
0.3194811147 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT22-like [Jatropha curcas] |
| 20 |
Hb_000951_060 |
0.3195666256 |
- |
- |
mads box protein, putative [Ricinus communis] |